8ORD
 
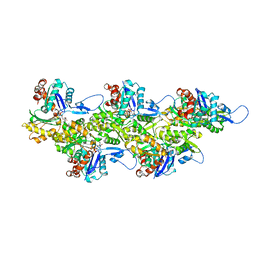 | | Cryo-EM map of zebrafish cardiac F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha 1b, ... | | Authors: | Bradshaw, M, Squire, J.M, Morris, E, Atkinson, G, Richardson, B, Lees, J, Paul, D.M. | | Deposit date: | 2023-04-13 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Zebrafish as a model for cardiac disease; Cryo-EM structure of native cardiac thin filaments from Danio Rerio.
J.Muscle Res.Cell.Motil., 44, 2023
|
|
8QYP
 
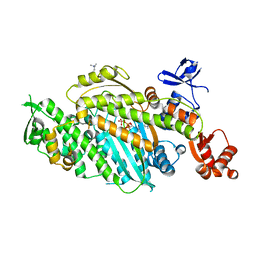 | | Beta-cardiac myosin motor domain in the pre-powerstroke state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin-7, ... | | Authors: | Robert-Paganin, J, Kikuti, C, Auguin, D, Rety, S, David, A, Houdusse, A. | | Deposit date: | 2023-10-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | Omecamtiv mecarbil and Mavacamten target the same myosin pocket despite antagonistic effects in heart contraction.
Biorxiv, 2023
|
|
8QYU
 
 | | Beta-cardiac myosin S1 fragment in the pre-powerstroke state complexed to Omecamtiv mecarbil | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Robert-Paganin, J, Kikuti, C, Auguin, D, Rety, S, David, A, Houdusse, A. | | Deposit date: | 2023-10-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Omecamtiv mecarbil and Mavacamten target the same myosin pocket despite antagonistic effects in heart contraction.
Biorxiv, 2023
|
|
7AP3
 
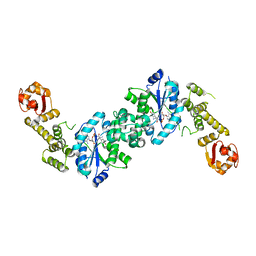 | | Crystal structure of E. coli tyrosyl-tRNA synthetase in complex with TyrS7HMDDA | | Descriptor: | SODIUM ION, Tyrosine--tRNA ligase, [(2~{R},3~{S},4~{R},5~{R})-5-[7-azanyl-5-(hydroxymethyl)benzimidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(2~{S})-2-azanyl-3-(4-hydroxyphenyl)propanoyl]sulfamate | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and Biological Evaluation of 1,3-Dideazapurine-Like 7-Amino-5-Hydroxymethyl-Benzimidazole Ribonucleoside Analogues as Aminoacyl-tRNA Synthetase Inhibitors.
Molecules, 25, 2020
|
|
8QYR
 
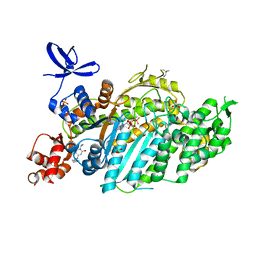 | | Beta-cardiac myosin motor domain in the pre-powerstroke state complexed to Mavacamten | | Descriptor: | 1,2-ETHANEDIOL, 6-[[(1~{S})-1-phenylethyl]amino]-3-propan-2-yl-1~{H}-pyrimidine-2,4-dione, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Robert-Paganin, J, Kikuti, C, Auguin, D, Rety, S, David, A, Houdusse, A. | | Deposit date: | 2023-10-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Omecamtiv mecarbil and Mavacamten target the same myosin pocket despite antagonistic effects in heart contraction.
Biorxiv, 2023
|
|
6V1D
 
 | | Crystal structure of human trefoil factor 1 | | Descriptor: | Trefoil factor 1 | | Authors: | Jarva, M.A, Lingford, J.P, John, A, Scott, N.E, Goddard-Borger, E.D. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Trefoil factors share a lectin activity that defines their role in mucus.
Nat Commun, 11, 2020
|
|
7AY3
 
 | |
2JMF
 
 | | Solution structure of the Su(dx) WW4- Notch PY peptide complex | | Descriptor: | E3 ubiquitin-protein ligase suppressor of deltex, Neurogenic locus Notch protein | | Authors: | Avis, J.M, Blankley, R.T, Jennings, M.D, Golovanov, A.P. | | Deposit date: | 2006-11-05 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and Autoregulation of Notch Binding by Tandem WW Domains in Suppressor of Deltex
J.Biol.Chem., 282, 2007
|
|
2JP0
 
 | | Solution structure of the N-terminal extraceullular domain of the lymphocyte receptor CD5 calculated using inferential structure determination (ISD) | | Descriptor: | T-cell surface glycoprotein CD5 | | Authors: | Garza-Garcia, A, Harris, R, Esposito, D, Driscoll, P.C, Rieping, W. | | Deposit date: | 2007-04-16 | | Release date: | 2008-02-26 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and conformational plasticity of the N-terminal scavenger receptor cysteine-rich domain of human CD5
J.Mol.Biol., 378, 2008
|
|
7AMD
 
 | | In situ assembly of choline acetyltransferase ligands by a hydrothiolation reaction reveals key determinants for inhibitor design | | Descriptor: | Choline O-acetyltransferase, SODIUM ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-2,2-dimethyl-4-[[3-[2-[(1~{R})-2-(1-methylpyridin-4-yl)-1-naphthalen-1-yl-ethyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Allgardsson, A, Ekstrom, F.J, Wiktelius, D, Bergstrom, T, Hoster, N, Akfur, C, Forsgren, N, Lejon, C, Hedenstrom, M, Linusson, A. | | Deposit date: | 2020-10-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | In Situ Assembly of Choline Acetyltransferase Ligands by a Hydrothiolation Reaction Reveals Key Determinants for Inhibitor Design.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2JN5
 
 | |
3E2V
 
 | | Crystal structure of an uncharacterized amidohydrolase from Saccharomyces cerevisiae | | Descriptor: | 3'-5'-exonuclease, GLYCEROL, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-06 | | Release date: | 2008-08-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized amidohydrolase from Saccharomyces cerevisiae
To be Published
|
|
8K76
 
 | |
2JOP
 
 | |
2JQH
 
 | | VPS4B MIT | | Descriptor: | Vacuolar protein sorting-associating protein 4B | | Authors: | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
2JQW
 
 | |
2JMR
 
 | | NMR structure of the E. coli type 1 pilus subunit FimF | | Descriptor: | fimF | | Authors: | Gossert, A.D, Bettendorff, P, Puorger, C, Vetsch, M, Herrmann, T, Fiorito, F, Hiller, S, Glockshuber, R, Wuthrich, K. | | Deposit date: | 2006-11-29 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Escherichia coli type 1 pilus subunit FimF and its interactions with other pilus subunits.
J.Mol.Biol., 375, 2008
|
|
2JNJ
 
 | | Solution structure of the p8 TFIIH subunit | | Descriptor: | TFIIH basal transcription factor complex TTD-A subunit | | Authors: | Vitorino, M, Atkinson, R.A, Moras, D, Poterszman, A, Kieffer, B, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2007-01-26 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Self-association Properties of the p8 TFIIH Subunit Responsible for Trichothiodystrophy
J.Mol.Biol., 368, 2007
|
|
2JNI
 
 | | Spatial structure of antimicrobial peptide arenicin-2 in aqueous solution | | Descriptor: | Arenicin-2 | | Authors: | Ovchinnikova, T.V, Shenkarev, Z.O, Nadezhdin, K.D, Balandin, S.V, Zhmak, M.N, Kudelina, I.A, Finkina, E.I, Kokryakov, V.N, Arseniev, A.S. | | Deposit date: | 2007-01-25 | | Release date: | 2007-08-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Recombinant expression, synthesis, purification, and solution structure of arenicin
Biochem.Biophys.Res.Commun., 360, 2007
|
|
2LU4
 
 | |
3EBA
 
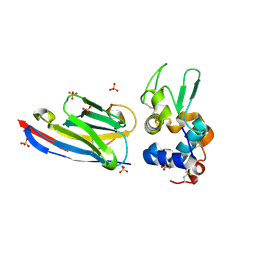 | | CAbHul6 FGLW mutant (humanized) in complex with human lysozyme | | Descriptor: | CAbHul6, Lysozyme C, SULFATE ION | | Authors: | Loris, R, Vincke, C, Saerens, D, Martinez-Rodriguez, S, Muyldermans, S, Conrath, K. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | General Strategy to Humanize a Camelid Single-domain Antibody and Identification of a Universal Humanized Nanobody Scaffold
J.Biol.Chem., 284, 2009
|
|
8QLO
 
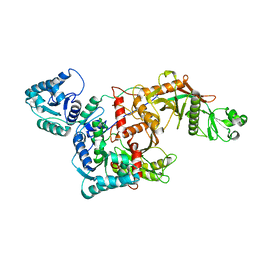 | | CryoEM structure of the apo SPARTA (BabAgo/TIR-APAZ) complex | | Descriptor: | Short prokaryotic Argonaute, Toll/interleukin-1 receptor domain-containing protein | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
8URG
 
 | | Human mitochondrial calcium uniporter crystal structure (residues 74-165 resolved) with lithium | | Descriptor: | Calcium uniporter protein, mitochondrial | | Authors: | Grainger, R, Colussi, D.C, Noble, M, Junop, M, Stathopulos, P.B. | | Deposit date: | 2023-10-25 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Disrupting the network of co-evolving amino terminal domain residues relieves mitochondrial calcium uptake inhibition by MCUb.
Comput Struct Biotechnol J, 27, 2025
|
|
2LVH
 
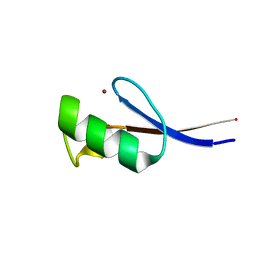 | | Solution structure of the zinc finger AFV1p06 protein from the hyperthermophilic archaeal virus AFV1 | | Descriptor: | Putative zinc finger protein ORF59a, ZINC ION | | Authors: | Guilliere, F, Sezonov, G, Prangishvili, D, Delepierre, M, Guijarro, J. | | Deposit date: | 2012-07-05 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an archaeal DNA binding protein with an eukaryotic zinc finger fold.
Plos One, 8, 2013
|
|
8VHV
 
 | |
