1MXL
 
 | |
1N87
 
 | |
1NA4
 
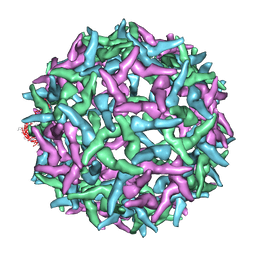 | | The structure of immature Yellow Fever virus particle | | Descriptor: | major envelope protein E | | Authors: | Zhang, Y, Corver, J, Chipman, P.R, Lenches, E, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-11-26 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of immature flavivirus particles
EMBO J., 22, 2003
|
|
4BKA
 
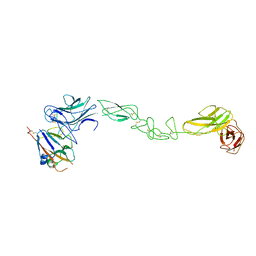 | | crystal structure of the human EphA4 ectodomain in complex with human ephrin A5 | | Descriptor: | EPHRIN TYPE-A RECEPTOR 4, EPHRIN-A5 | | Authors: | Seiradake, E, Schaupp, A, del Toro Ruiz, D, Kaufmann, R, Mitakidis, N, Harlos, K, Aricescu, A.R, Klein, R, Jones, E.Y. | | Deposit date: | 2013-04-23 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (5.3 Å) | | Cite: | Structurally Encoded Intraclass Differences in Epha Clusters Drive Distinct Cell Responses
Nat.Struct.Mol.Biol., 20, 2013
|
|
4BLF
 
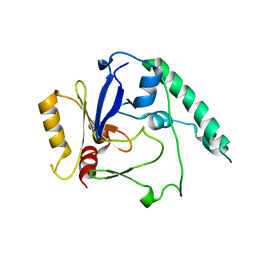 | | Variable internal flexibility characterizes the helical capsid formed by Agrobacterium VirE2 protein on single-stranded DNA. | | Descriptor: | SINGLE-STRAND DNA-BINDING PROTEIN | | Authors: | Bharat, T.A.M, Zbaida, D, Eisenstein, M, Frankenstein, Z, Mehlman, T, Weiner, L, Sorzano, C.O.S, Barak, Y, Albeck, S, Briggs, J.A.G, Wolf, S.G, Elbaum, M. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Variable Internal Flexibility Characterizes the Helical Capsid Formed by Agrobacterium Vire2 Protein on Single-Stranded DNA.
Structure, 21, 2013
|
|
1LUM
 
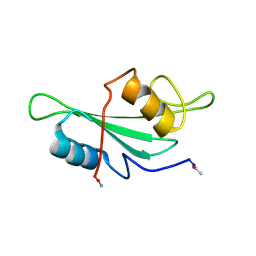 | | NMR Structure of the Itk SH2 domain, Pro287trans, 20 low energy structures | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Mallis, R.J, Brazin, K.N, Fulton, D.B, Andreotti, A.H. | | Deposit date: | 2002-05-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of a proline-driven conformational switch
within the Itk SH2 domain
Nat.Struct.Biol., 9, 2002
|
|
1LX8
 
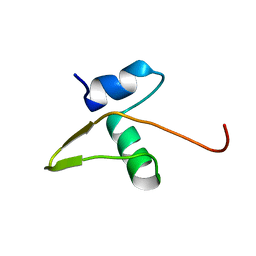 | | Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein | | Descriptor: | Excisionase | | Authors: | Sam, M.D, Papagiannis, C, Connolly, K.M, Corselli, L, Iwahara, J, Lee, J, Phillips, M, Wojciak, J.M, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2002-06-04 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein
J.Mol.Biol., 324, 2002
|
|
1N6G
 
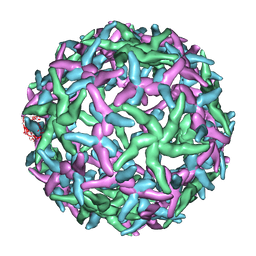 | | The structure of immature Dengue-2 prM particles | | Descriptor: | major envelope protein E | | Authors: | Zhang, Y, Corver, J, Chipman, P.R, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-11-10 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structures of Immature flavivirus particles
EMBO J., 22, 2003
|
|
1N8X
 
 | | Solution structure of HIV-1 Stem Loop SL1 | | Descriptor: | HIV-1 STEM LOOP SL1 MONOMERIC RNA | | Authors: | Lawrence, D.C, Stover, C.C, Noznitsky, J, Wu, Z, Summers, M.F. | | Deposit date: | 2002-11-21 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Intact Stem and Bulge of HIV-1 Psi-RNA Stem-Loop SL1
J.Mol.Biol., 326, 2003
|
|
1NHA
 
 | | Solution Structure of the Carboxyl-Terminal Domain of RAP74 and NMR Characterization of the FCP-Binding Sites of RAP74 and CTD of RAP74, the subunit of Human TFIIF | | Descriptor: | Transcription initiation factor IIF, alpha subunit | | Authors: | Nguyen, B.D, Chen, H.T, Kobor, M.S, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Carboxyl-Terminal Domain of RAP74 and NMR Characterization of the FCP1-Binding Sites of RAP74 and Human TFIIB.
Biochemistry, 42, 2003
|
|
1NA2
 
 | |
1NIY
 
 | |
7OFS
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 4-(2-hydroxyethyl)phenol | | Descriptor: | 4-(2-hydroxyethyl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Ewert, W, Sprenger, J, Koua, F, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Wolf, M, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFT
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with p-hydroxybenzaldehyde | | Descriptor: | CHLORIDE ION, P-HYDROXYBENZALDEHYDE, POTASSIUM ION, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Ewert, W, Sprenger, J, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Wolf, M, Yefanov, O, Gelisio, L, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7ZPG
 
 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH LIGAND | | Descriptor: | Monoglyceride lipase, [(7R,9aR)-7-(4-chlorophenyl)-1,3,4,6,7,8,9,9a-octahydropyrido[1,2-a]pyrazin-2-yl]-(2-bromanyl-3-methoxy-phenyl)methanone | | Authors: | Kemble, A, Hornsperger, B, Ruf, I, Richter, H, Benz, J, Kuhn, B, Heer, D, Wittwer, M, Engelhardt, B, Grether, U, Collin, L, Leibrock, L. | | Deposit date: | 2022-04-27 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | A potent and selective inhibitor for the modulation of MAGL activity in the neurovasculature.
Plos One, 17, 2022
|
|
7ZTS
 
 | | Saccharomyces cerevisiae L-BC virus, open particle, asymmetric reconstruction | | Descriptor: | Major capsid protein | | Authors: | Grybchuk, D, Prochazkova, M, Fuzik, T, Konovalovas, A, Serva, S, Yurchenko, V, Plevka, P. | | Deposit date: | 2022-05-11 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structures of L-BC virus and its open particle provide insight into Totivirus capsid assembly.
Commun Biol, 5, 2022
|
|
1CA3
 
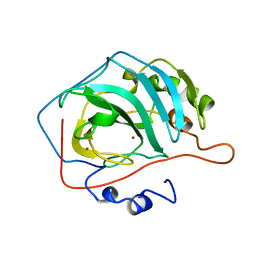 | |
1C80
 
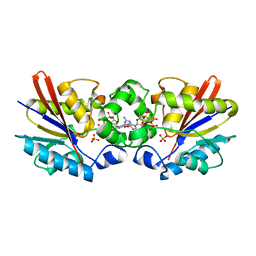 | | REGULATORY COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
1C7Z
 
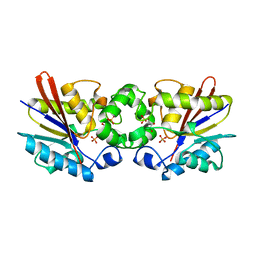 | | REGULATORY COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, GLYCERALDEHYDE-3-PHOSPHATE, PHOSPHATE ION | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
7PTV
 
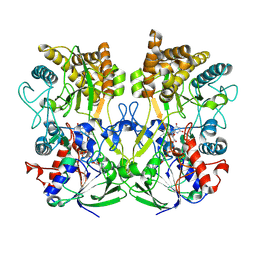 | | Structure of the Mimivirus genomic fibre asymmetric unit | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | Descriptor: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | Deposit date: | 2018-12-15 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
6NZS
 
 | | Dextranase AoDex KQ11 | | Descriptor: | Dextranase | | Authors: | Ren, W, Yan, W, Gu, L, Feng, Y, Dong, D, Wang, S, Wang, C, Lyu, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
To Be Published
|
|
5KZL
 
 | | Structure of Heme Oxygenase from Leptospira interrogans | | Descriptor: | Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Klinke, S, Soldano, A, Otero, L.H, Rivera, M, Catalano-Dupuy, D.L, Ceccarelli, E.A. | | Deposit date: | 2016-07-25 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and mutational analyses of the Leptospira interrogans virulence-related heme oxygenase provide insights into its catalytic mechanism.
PLoS ONE, 12, 2017
|
|
7ZQ4
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ and the oxidated catalytic cysteine | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase A, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
7OMA
 
 | | Thosea asigna virus RdRP domain elongation complex | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, RNA (5'-R(P*AP*AP*AP*UP*UP*UP*U)-3'), ... | | Authors: | Ferrero, D.S, Falqui, M, Verdaguer, N. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of a Non-Canonical RdRP in Action.
Viruses, 13, 2021
|
|
