3EYM
 
 | | Structure of Influenza Haemagglutinin in complex with an inhibitor of membrane fusion | | Descriptor: | 2-tert-butylbenzene-1,4-diol, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Russell, R.J, Kerry, P.S, Stevens, D.A, Steinhauer, D.A, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2008-10-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4S14
 
 | | Crystal structure of the orphan nuclear receptor RORgamma ligand-binding domain in complex with 4alpha-caboxyl, 4beta-methyl-zymosterol (4ACD8) | | Descriptor: | (3beta,4alpha,5beta,14beta)-3-hydroxy-4-methylcholesta-8,24-diene-4-carboxylic acid, Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1 | | Authors: | Huang, P, Santori, F.R, Littman, D.R, Rastinejad, F. | | Deposit date: | 2015-01-07 | | Release date: | 2015-02-11 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (3.542 Å) | | Cite: | Identification of Natural ROR gamma Ligands that Regulate the Development of Lymphoid Cells.
Cell Metab, 21, 2015
|
|
7SVM
 
 | | DPP8 IN COMPLEX WITH LIGAND ICeD-2 | | Descriptor: | (2S)-2-amino-1-(1,3-dihydro-2H-isoindol-2-yl)-2-[(1r,4S)-4-(pyrrolidin-1-yl)cyclohexyl]ethan-1-one, Dipeptidyl peptidase 8, trimethylamine oxide | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVN
 
 | | DPP9 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 9 | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVO
 
 | | DPP8 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 8, trimethylamine oxide | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
8A1E
 
 | | Rabies virus glycoprotein in complex with Fab fragments of 17C7 and 1112-1 neutralizing antibodies | | Descriptor: | Fab 1112-1 heavy chain variable domain, Fab 1112-1 light chain variable domain, Fab 17C7 heavy chain variable domain, ... | | Authors: | Ng, W.M, Fedosyuk, S, English, S, Augusto, G, Berg, A, Thorley, L, Haselon, A.S, Segireddy, R.R, Bowden, T.A, Douglas, A.D. | | Deposit date: | 2022-06-01 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure of trimeric pre-fusion rabies virus glycoprotein in complex with two protective antibodies.
Cell Host Microbe, 30, 2022
|
|
5K3Q
 
 | | Rhesus macaques Trim5alpha Bbox2 domain | | Descriptor: | NITRATE ION, Tripartite motif-containing protein 5,Tripartite motif-containing protein 5, ZINC ION | | Authors: | Keown, J.K, Goldstone, D.C. | | Deposit date: | 2016-05-19 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Trim5 alpha Bbox2 domain from rhesus macaques describes a plastic oligomerisation interface.
J.Struct.Biol., 195, 2016
|
|
5CRL
 
 | |
5K4B
 
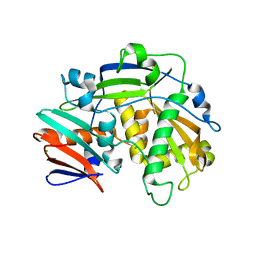 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 1 | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit D | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
6VIU
 
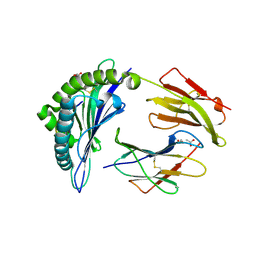 | |
4UV3
 
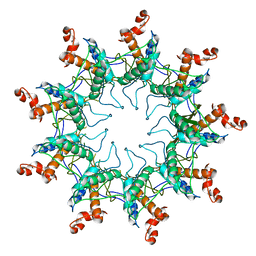 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
6PE5
 
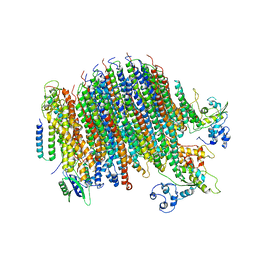 | | Yeast Vo motor in complex with 2 VopQ molecules | | Descriptor: | Cation transporter, Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, ... | | Authors: | Peng, W, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2019-06-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A distinct inhibitory mechanism of the V-ATPase by Vibrio VopQ revealed by cryo-EM.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PFP
 
 | | Crystal Structure of Amino Acids 1473-1536 of Human Beta Cardiac Myosin Fused to Gp7 and Eb1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Myosin-7 fused to GP7 and EB1, SULFATE ION | | Authors: | Andreas, M.P, Korkmaz, E.N, Kirsch, C.J, Hargreaves, M.D, Kieffer, D.J, Ajay, G, Cui, Q, Rayment, I. | | Deposit date: | 2019-06-21 | | Release date: | 2020-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Complete Model of the Cardiac Myosin Rod
To Be Published
|
|
7AWE
 
 | | HUMAN IMMUNOPROTEASOME 20S PARTICLE IN COMPLEX WITH [(1R)-2-(1-benzofuran-3-yl)-1-{[(1S,2R,4R)-7-oxabicyclo[2.2.1]heptan-2-yl]formamido}ethyl]boronic acid | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Musil, D, Klein, M. | | Deposit date: | 2020-11-06 | | Release date: | 2021-06-09 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | M3258 Is a Selective Inhibitor of the Immunoproteasome Subunit LMP7 ( beta 5i) Delivering Efficacy in Multiple Myeloma Models.
Mol.Cancer Ther., 20, 2021
|
|
3E3O
 
 | | Glycogen phosphorylase R state-IMP complex | | Descriptor: | Glycogen phosphorylase, muscle form, INOSINIC ACID, ... | | Authors: | Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-08-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Glycogen phosphorylase revisited: extending the resolution of the R- and T-state structures of the free enzyme and in complex with allosteric activators.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
5DCW
 
 | | Iridoid synthase from Catharanthus roseus - ligand free structure | | Descriptor: | 1,2-ETHANEDIOL, Iridoid synthase | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
5DD8
 
 | | The Crystal structure of HucR mutant (HucR-E48Q) from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, Transcriptional regulator, MarR family | | Authors: | Deochand, D.K, Perera, I.C, Crochet, R.B, Gilbert, N.C, Newcomer, M.E, Grove, A. | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Histidine switch controlling pH-dependent protein folding and DNA binding in a transcription factor at the core of synthetic network devices.
Mol Biosyst, 12, 2016
|
|
7ZJ3
 
 | | Structure of TRIM2 RING domain in complex with UBE2D1~Ub conjugate | | Descriptor: | Polyubiquitin-C, Tripartite motif-containing protein 2, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Esposito, D, Garza-Garcia, A, Dudley-Fraser, J, Rittinger, K. | | Deposit date: | 2022-04-08 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Divergent self-association properties of paralogous proteins TRIM2 and TRIM3 regulate their E3 ligase activity.
Nat Commun, 13, 2022
|
|
6ICD
 
 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
4W6A
 
 | |
5D11
 
 | | Kinase domain of cSrc in complex with RL235 | | Descriptor: | GLYCEROL, N-[3-({4-(4-methylpiperazin-1-yl)-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl}oxy)phenyl]prop-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Becker, C, Gruetter, C, Engel, J, Rauh, D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting Drug Resistance in EGFR with Covalent Inhibitors: A Structure-Based Design Approach.
J.Med.Chem., 58, 2015
|
|
5JQX
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with phosphoglyceric acid (PGA) - GpgS*PGA | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Glucosyl-3-phosphoglycerate synthase | | Authors: | Albesa-Jove, D, Sancho-Vaello, E, Rodrigo-Unzueta, A, Comino, N, Carreras-Gonzalez, A, Arrasate, P, Urresti, S, Guerin, M.E. | | Deposit date: | 2016-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Snapshots and Loop Dynamics along the Catalytic Cycle of Glycosyltransferase GpgS.
Structure, 25, 2017
|
|
2VHI
 
 | | Crystal structure of a pyrimidine degrading enzyme from Drosophila melanogaster | | Descriptor: | CG3027-PA | | Authors: | Lundgren, S, Lohkamp, B, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-11-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of Beta-Alanine Synthase from Drosophila Melanogaster Reveals a Homooctameric Helical Turn-Like Assembly.
J.Mol.Biol., 377, 2008
|
|
1JTZ
 
 | | CRYSTAL STRUCTURE OF TRANCE/RANKL CYTOKINE. | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 11 | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2001-08-23 | | Release date: | 2001-09-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the TRANCE/RANKL cytokine reveals determinants of receptor-ligand specificity
J.Clin.Invest., 108, 2001
|
|
1JYD
 
 | | Crystal Structure of Recombinant Human Serum Retinol-Binding Protein at 1.7 A Resolution | | Descriptor: | GLYCEROL, PLASMA RETINOL-BINDING PROTEIN | | Authors: | Greene, L.H, Chrysina, E.D, Irons, L.I, Papageorgiou, A.C, Acharya, K.R, Brew, K. | | Deposit date: | 2001-09-12 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of conserved residues in structure and stability: Tryptophans of human serum
retinol-binding protein, a model for the lipocalin superfamily
Protein Sci., 10, 2001
|
|
