7S5I
 
 | | Crystal structure of Aldose-6-phosphate reductase (Ald6PRase) from peach (Prunus persica) leaves | | Descriptor: | Sorbitol-6-phosphate dehydrogenase | | Authors: | Zheng, Y, Bhayani, J.A, Romina, I.M, Hartman, M.D, Cereijo, A.E, Ballicora, M.A, Iglesias, A.A, Figueroa, C.M, Liu, D. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural Determinants of Sugar Alcohol Biosynthesis in Plants: The Crystal Structures of Mannose-6-Phosphate and Aldose-6-Phosphate Reductases.
Plant Cell.Physiol., 63, 2022
|
|
1BP1
 
 | | CRYSTAL STRUCTURE OF BPI, THE HUMAN BACTERICIDAL PERMEABILITY-INCREASING PROTEIN | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERICIDAL/PERMEABILITY-INCREASING PROTEIN | | Authors: | Beamer, L.J, Carroll, S.F, Eisenberg, D. | | Deposit date: | 1997-04-08 | | Release date: | 1997-09-04 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution.
Science, 276, 1997
|
|
7Z3H
 
 | | Crystal structure of Iba57 from Chaetomium thermophilum | | Descriptor: | CITRIC ACID, Transferase-like protein | | Authors: | Altegoer, F, Mrusek, D, Bange, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The iron-sulfur cluster assembly (ISC) protein Iba57 executes a tetrahydrofolate-independent function in mitochondrial [4Fe-4S] protein maturation.
J.Biol.Chem., 298, 2022
|
|
8FLT
 
 | | Human PTH1R in complex with M-PTH(1-14) and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cary, B.P, Belousoff, M.J, Piper, S.J, Wootten, D, Sexton, P.M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-26 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Molecular insights into peptide agonist engagement with the PTH receptor.
Structure, 31, 2023
|
|
7UUL
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with kanamycin B and coenzyme A | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
6VWL
 
 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P/E tRNA (rotated conformation) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
7O9J
 
 | | Crystal structure of DyP-type peroxidase from Dictyostelium discoideum in complex with an activated form of oxygen | | Descriptor: | 1,2-ETHANEDIOL, DyPA, OXYGEN MOLECULE, ... | | Authors: | Rai, A, Fedorov, R, Manstein, D.J. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of a Dye-Decolorizing Peroxidase from Dictyostelium discoideum .
Int J Mol Sci, 22, 2021
|
|
4GX1
 
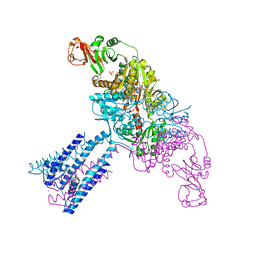 | | Crystal structure of the GsuK bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | Deposit date: | 2012-09-03 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
7O9L
 
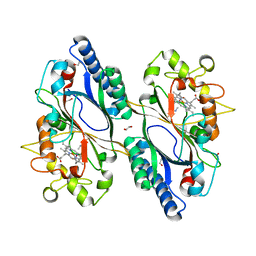 | | Dictyostelium discoideum dye decolorizing peroxidase DyPA in complex with cyanide. | | Descriptor: | 1,2-ETHANEDIOL, CYANIDE ION, DyPA, ... | | Authors: | Rai, A, Fedorov, R, Manstein, D.J. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of a Dye-Decolorizing Peroxidase from Dictyostelium discoideum .
Int J Mol Sci, 22, 2021
|
|
6VZ6
 
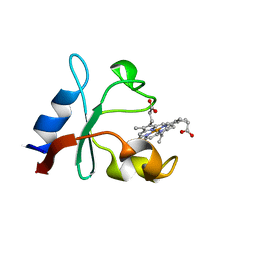 | | Methanococcoides burtonii cytochrome b5 domain protein (WP 011499504.1) | | Descriptor: | Cytochrome b5-domain protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Teakel, S.L, Forwood, J.K, Aragao, D, Cahill, M.A, Marama, M. | | Deposit date: | 2020-02-27 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methanococcoides burtonii cytochrome b5 domain protein (WP 011499504.1)
To Be Published
|
|
8IHR
 
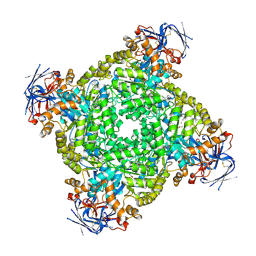 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | Descriptor: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
4GSX
 
 | | High resolution structure of dengue virus serotype 1 sE containing stem | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Klein, D.E, Choi, J.L, Harrison, S.C. | | Deposit date: | 2012-08-28 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structure of a dengue virus envelope protein late-stage fusion intermediate.
J.Virol., 87, 2013
|
|
6NDD
 
 | |
8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | Descriptor: | Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
6R8N
 
 | | STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Colletier, J.-P, Gauto, D, Estrozi, L, Favier, A, Effantin, G, Schoehn, G, Boisbouvier, J, Schanda, P. | | Deposit date: | 2019-04-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
4Z7M
 
 | | Novel Inhibitors of Bacterial Methionine Aminopeptidase with Broad-Spectrum Biochemical Activity | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase, N~2~-[(3,5-difluorophenyl)acetyl]-N-[(3S,7R)-1-methyl-2-oxo-7-phenyl-2,3,4,7-tetrahydro-1H-azepin-3-yl]-L-alaninamide | | Authors: | Rose, J.A, Lahiri, S.D, McKinney, D.C, Albert, R, Morningstar, M, Shapiro, A.B, Fisher, S.F, Fleming, P.R. | | Deposit date: | 2015-04-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Novel Inhibitors of Bacterial Methionine Aminopeptidase with Broad-Spectrum Biochemical Activity
To be Published
|
|
5KVE
 
 | | Zika specific antibody, ZV-48, bound to ZIKA envelope DIII | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Genome polyprotein, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
3NHA
 
 | | Nucleotide Binding Domain of Human ABCB6 (ADP Mg bound structure) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-binding cassette sub-family B member 6, mitochondrial, ... | | Authors: | Haffke, M, Menzel, A, Carius, Y, Jahn, D, Heinz, D.W. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6QOZ
 
 | | CryoEM reconstruction of Cowpea Mosaic Virus (CPMV) bound to an Affimer reagent | | Descriptor: | Affimer binding protein, Cowpea mosaic virus large subunit, RNA2 polyprotein | | Authors: | Hesketh, E.L, Tiede, C, Adamson, H, Adams, T.L, Byrne, M.J, Meshcheriakova, Y, Lomonossoff, G.P, Kruse, I, McPherson, M.J, Tomlinson, D.C, Ranson, N.A. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Affimer reagents as tools in diagnosing plant virus diseases.
Sci Rep, 9, 2019
|
|
1BUX
 
 | | 3'-PHOSPHORYLATED NUCLEOTIDES BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Xu, Y, Schneider, B, Deville-Bonne, D, Veron, M, Janin, J. | | Deposit date: | 1998-09-07 | | Release date: | 1999-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3'-Phosphorylated nucleotides are tight binding inhibitors of nucleoside diphosphate kinase activity.
J.Biol.Chem., 273, 1998
|
|
5MBE
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
1BN3
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | 2-(3-METHOXYPHENYL)-2H-THIENO-[3,2-E]-1,2-THIAZINE-6-SULFINAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-31 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BNN
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | 3,,4-DIHYDRO-2-(3-METHOXYPHENYL)-2H-THIENO-[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
2X93
 
 | | Crystal structure of AnCE-trandolaprilat complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ANGIOTENSIN CONVERTING ENZYME, ... | | Authors: | Akif, M, Georgiadis, D, Mahajan, A, Dive, V, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High Resolution Crystal Structures of Drosophila Melanogaster Angiotensin Converting Enzyme in Complex with Novel Inhibitors and Anti- Hypertensive Drugs.
J.Mol.Biol., 400, 2010
|
|
1BU5
 
 | | X-RAY CRYSTAL STRUCTURE OF THE DESULFOVIBRIO VULGARIS (HILDENBOROUGH) APOFLAVODOXIN-RIBOFLAVIN COMPLEX | | Descriptor: | PROTEIN (FLAVODOXIN), RIBOFLAVIN, SULFATE ION | | Authors: | Walsh, M.A, Mccarthy, A, O'Farrell, P.A, Mccardle, P, Cunningham, P.D, Mayhew, S.G, Higgins, T.M. | | Deposit date: | 1998-09-12 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray crystal structure of the Desulfovibrio vulgaris (Hildenborough) apoflavodoxin-riboflavin complex.
Eur.J.Biochem., 258, 1998
|
|
