5IQJ
 
 | | 1.9 Angstrom Crystal Structure of Protein with Unknown Function from Vibrio cholerae. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Skarina, T, Seed, K.D, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Protein with Unknown Function from Vibrio cholerae.
To Be Published
|
|
6V0L
 
 | | PDGFR-b Promoter Forms a G-Vacancy Quadruplex that Can be Complemented by dGMP: Molecular Structure and Recognition of Guanine Derivatives and Metabolites | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*(3D1)P*AP*GP*GP*GP*AP*GP*GP*GP*CP*GP*GP*CP*GP*GP*GP*AP*CP*A)-3') | | Authors: | Wang, K.B, Dickerhoff, J, Wu, G, Yang, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PDGFR-beta Promoter Forms a Vacancy G-Quadruplex that Can Be Filled in by dGMP: Solution Structure and Molecular Recognition of Guanine Metabolites and Drugs.
J.Am.Chem.Soc., 142, 2020
|
|
6PQ3
 
 | | Crystal structure of GDP-bound KRAS with ten residues long internal tandem duplication in the switch II region | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dharmaiah, S, Chan, A.H, Tran, T.H, Simanshu, D.K. | | Deposit date: | 2019-07-08 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | RASinternal tandem duplication disrupts GTPase-activating protein (GAP) binding to activate oncogenic signaling.
J.Biol.Chem., 295, 2020
|
|
4Y4O
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome with rRNA modifications and bound to protein Y (YfiA) at 2.3A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Polikanov, Y.S, Melnikov, S.V, Soll, D, Steitz, T.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8B51
 
 | | Usutu virus methyltransferase domain in complex with sinefungin | | Descriptor: | GLYCEROL, GLYCINE, RNA-directed RNA polymerase NS5, ... | | Authors: | Ferrero, D.S, Albentosa Gonzalez, L, Mas, A, Verdaguer, N. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure and function of the NS5 methyltransferase domain from Usutu virus.
Antiviral Res., 208, 2022
|
|
6YM5
 
 | | Crystal structure of BAY-091 with PIP4K2A | | Descriptor: | (2~{R})-2-[[3-cyano-2-[4-(2-fluoranyl-3-methyl-phenyl)phenyl]-1,7-naphthyridin-4-yl]amino]butanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
4YAH
 
 | | Crystal Structure of the Methionine Binding Protein, MetQ | | Descriptor: | D-methionine-binding lipoprotein MetQ, METHIONINE | | Authors: | Lai, J.Y, Kadaba, N.S, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2015-02-17 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The contribution of methionine to the stability of the Escherichia coli MetNIQ ABC transporter-substrate binding protein complex.
Biol.Chem., 396, 2015
|
|
4A9V
 
 | | Pseudomonas fluorescens PhoX | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2011-11-28 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
8F2L
 
 | |
8B52
 
 | | Usutu virus methyltransferase domain in complex with sinefungin | | Descriptor: | GLYCEROL, Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Ferrero, D.S, Albentosa Gonzalez, L, Mas, A, Verdaguer, N. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and function of the NS5 methyltransferase domain from Usutu virus.
Antiviral Res., 208, 2022
|
|
6N6K
 
 | | Human REXO2 bound to pAG | | Descriptor: | MALONATE ION, RNA (5'-R(P*AP*G)-3'), RNA exonuclease 2 homolog,Small fragment nuclease, ... | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
5IT3
 
 | | Swirm domain of human Lsd1 | | Descriptor: | Lysine-specific histone demethylase 1A, MAGNESIUM ION | | Authors: | Jeffrey, P.D, Yuan, P. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Tlx-interacting peptide of Lsd1 inhibits the proliferation of brain tumor stem cells
To Be Published
|
|
8H0N
 
 | | Crystal structure of the human METTL1-WDR4 complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 | | Authors: | Jin, X.H, Guan, Z.Y, Gong, Z, Zhang, D.L. | | Deposit date: | 2022-09-30 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into how WDR4 promotes the tRNA N7-methylguanosine methyltransferase activity of METTL1.
Cell Discov, 9, 2023
|
|
8P3Q
 
 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
6MNC
 
 | | CRYSTAL STRUCTURE OF HUMAN 17BETA-HYDROXYSTEROID DEHYDROGENASE TYPE 1 COMPLEXED WITH ESTRONE | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, DI(HYDROXYETHYL)ETHER, Estradiol 17-beta-dehydrogenase 1 | | Authors: | Li, T, Stephen, P, Zhu, D.W, Lin, S.X. | | Deposit date: | 2018-10-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of human 17 beta-hydroxysteroid dehydrogenase type 1 complexed with estrone and NADP+reveal the mechanism of substrate inhibition.
Febs J., 286, 2019
|
|
6J2M
 
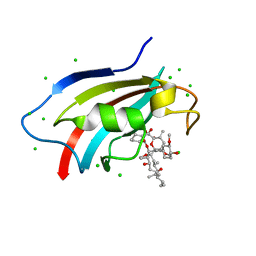 | | Crystal structure of AtFKBP53 C-terminal domain | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP53 | | Authors: | Singh, A.K, Vasudevan, D. | | Deposit date: | 2019-01-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | AtFKBP53: a chimeric histone chaperone with functional nucleoplasmin and PPIase domains.
Nucleic Acids Res., 48, 2020
|
|
8ENL
 
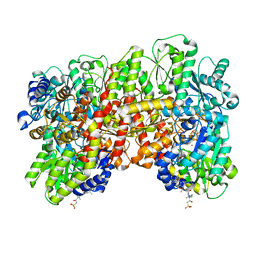 | | CryoEM structure of the high pH turnover-inactivated nitrogenase MoFe-protein | | Descriptor: | CHAPSO, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
4Y6U
 
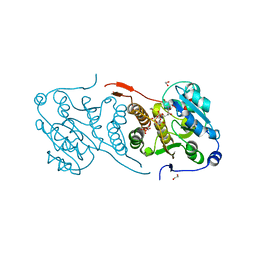 | | Mycobacterial protein | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, CHLORIDE ION, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-13 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8ENN
 
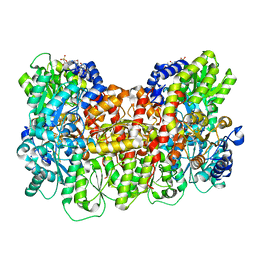 | | Homocitrate-deficient nitrogenase MoFe-protein from Azotobacter vinelandii nifV knockout | | Descriptor: | CHAPSO, CITRIC ACID, FE (III) ION, ... | | Authors: | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
8ENM
 
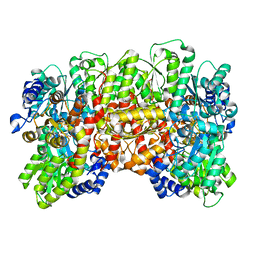 | | CryoEM structure of the high pH nitrogenase MoFe-protein under non-turnover conditions | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
8BLP
 
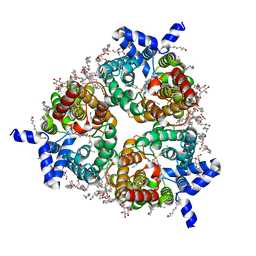 | | Human Urea Transporter UT-B/UT1 in Complex with Inhibitor UTBinh-14 | | Descriptor: | 10-(4-ethylphenyl)sulfonyl-~{N}-(thiophen-2-ylmethyl)-5-thia-1,8,11,12-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,7,9,11-pentaen-7-amine, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Chi, G, Dietz, L, Pike, A.C.W, Maclean, E.M, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, Scacioc, A, McKinley, G, Arrowsmith, C.H, Edwards, A, Bountra, C, Fernandez-Cid, A, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural characterization of human urea transporters UT-A and UT-B and their inhibition.
Sci Adv, 9, 2023
|
|
8ENO
 
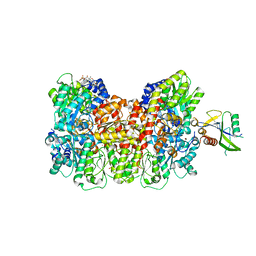 | | Homocitrate-deficient nitrogenase MoFe-protein from A. vinelandii nifV knockout in complex with NafT | | Descriptor: | CHAPSO, CITRIC ACID, FE (III) ION, ... | | Authors: | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
6MSX
 
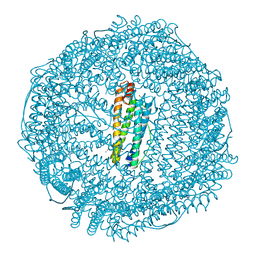 | | Iron containing ferritin at 1.43A | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Blackburn, A, Partowmah, S.H, Brennan, H.M, Mestizo, K.E, Stivala, C.D, Petreczky, J, Perez, A, Horn, A, McSweeney, S. | | Deposit date: | 2018-10-18 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A simple technique to improve microcrystals using gel exclusion of nucleation inducing elements
To Be Published
|
|
5JL9
 
 | |
6YRZ
 
 | | Crystal structure of FAP et pH 8.5 after illumination at 150K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, ... | | Authors: | Sorigue, D, Legrand, P, Blangy, S, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
