7SQI
 
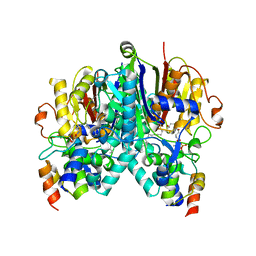 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C14-crypto Acyl Carrier Protein, AcpP | | Descriptor: | Acyl carrier protein, Beta-ketoacyl-ACP synthase I, N-{2-[(2Z)-3-chlorotetradec-2-enamido]ethyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Chen, A, Mindrebo, J.T, Davis, T.D, Noel, J.P, Burkart, M.D. | | Deposit date: | 2021-11-05 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5JUC
 
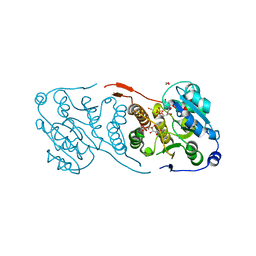 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate (UDP) and glucosyl-3-phosphoglycerate (GPG) - GpgS*GPG*UDP*Mn2+_2 | | Descriptor: | (2R)-2-(alpha-D-glucopyranosyloxy)-3-(phosphonooxy)propanoic acid, 1,2-ETHANEDIOL, Glucosyl-3-phosphoglycerate synthase, ... | | Authors: | Albesa-Jove, D, Sancho-Vaello, E, Rodrigo-Unzueta, A, Comino, N, Carreras-Gonzalez, A, Arrasate, P, Urresti, S, Guerin, M.E. | | Deposit date: | 2016-05-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Snapshots and Loop Dynamics along the Catalytic Cycle of Glycosyltransferase GpgS.
Structure, 25, 2017
|
|
5NOG
 
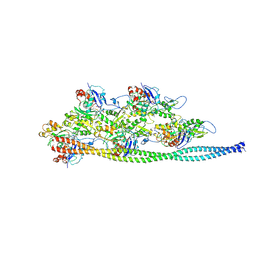 | | Ca2+-induced Movement of Tropomyosin on Native Cardiac Thin Filaments - "Blocked" state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cardiac muscle alpha actin 1, MAGNESIUM ION, ... | | Authors: | Risi, C, Eisner, J, Belknap, B, Heeley, D.H, White, H.D, Schroeder, G.F, Galkin, V.E. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-19 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Ca(2+)-induced movement of tropomyosin on native cardiac thin filaments revealed by cryoelectron microscopy.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8FD5
 
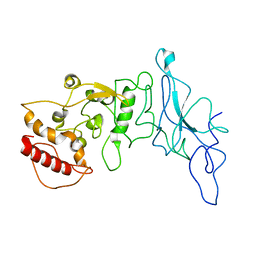 | | Nucleocapsid monomer structure from SARS-CoV-2 | | Descriptor: | Nucleoprotein | | Authors: | Casasanta, M, Jonaid, G.M, Kaylor, L, Luqiu, W, DiCecco, L, Solares, M, Berry, S, Kelly, D.F. | | Deposit date: | 2022-12-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (4.57 Å) | | Cite: | Structural Insights of the SARS-CoV-2 Nucleocapsid Protein: Implications for the Inner-workings of Rapid Antigen Tests.
Microsc Microanal, 29, 2023
|
|
6V6L
 
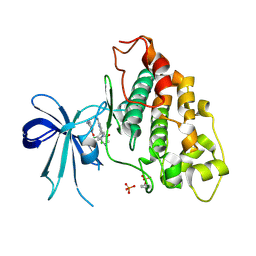 | | Co-structure of human glycogen synthase kinase beta with 1-(6-((2-((6-amino-5-nitropyridin-2-yl)amino)ethyl)amino)-2-(2,4-dichlorophenyl)pyridin-3-yl)-4-methylpiperazin-2-one | | Descriptor: | 1-(6-((2-((6-amino-5-nitropyridin-2-yl)amino)ethyl)amino)-2-(2,4-dichlorophenyl)pyridin-3-yl)-4-methylpiperazin-2-one, Glycogen synthase kinase-3 beta, PHOSPHATE ION | | Authors: | Bussiere, D.E, Fang, E, Shu, W. | | Deposit date: | 2019-12-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery and optimization of novel pyridines as highly potent and selective glycogen synthase kinase 3 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4X53
 
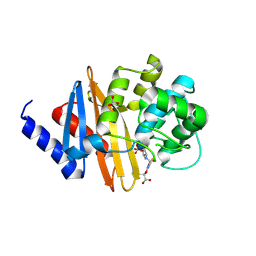 | | Structure of the class D Beta-Lactamase OXA-160 V130D in Acyl-Enzyme Complex with Aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, BICARBONATE ION, Class D beta-lactamase OXA-160 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
6P9B
 
 | |
4X56
 
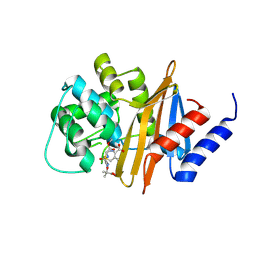 | | Structure of the class D Beta-Lactamase OXA-160 V130D in Acyl-Enzyme Complex with Ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Class D beta-lactamase OXA-160 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
7USA
 
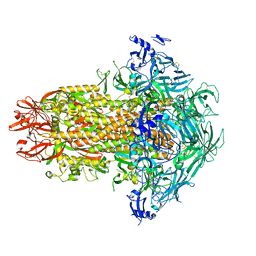 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
5KC4
 
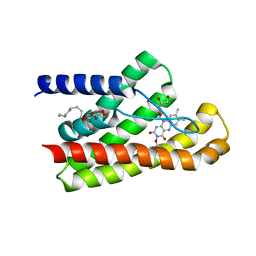 | | Structure of TmRibU, orthorhombic crystal form | | Descriptor: | RIBOFLAVIN, Riboflavin transporter RibU, nonyl beta-D-glucopyranoside | | Authors: | Karpowich, N.K, Wang, D.N, Song, J.M. | | Deposit date: | 2016-06-04 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Aromatic Cap Seals the Substrate Binding Site in an ECF-Type S Subunit for Riboflavin.
J.Mol.Biol., 428, 2016
|
|
4X5Z
 
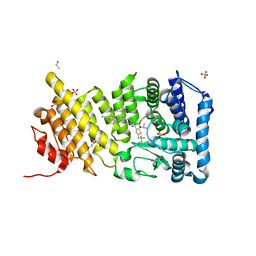 | | menin in complex with MI-136 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Borkin, D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pharmacologic Inhibition of the Menin-MLL Interaction Blocks Progression of MLL Leukemia In Vivo.
Cancer Cell, 27, 2015
|
|
7BH1
 
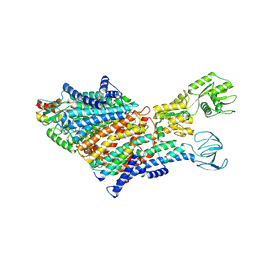 | | Cryo-EM Structure of KdpFABC in E1 state with K | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BGY
 
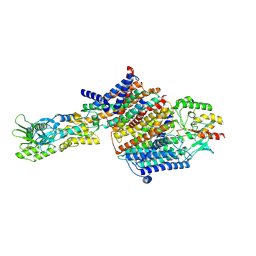 | | Cryo-EM Structure of KdpFABC in E2Pi state with MgF4 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XWN
 
 | |
6V9D
 
 | |
5JZ9
 
 | | Crystal structure of HsaD bound to 3,5-dichloro-4-hydroxybenzenesulphonic acid | | Descriptor: | 3,5-dichloro-4-hydroxybenzene-1-sulfonic acid, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
7T0U
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-12387 | | Descriptor: | 3-chloro-5-{7-[2-({5-chloro-2-[(2S)-2-methyl-4-(oxetan-3-yl)piperazin-1-yl]pyridin-4-yl}amino)-2-oxoethyl]-3-methyl-4-oxo-2-(trifluoromethyl)-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-hydroxybenzamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-11-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-12387
To Be Published
|
|
7NZN
 
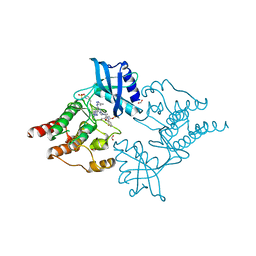 | | Structure of RET kinase domain bound to inhibitor JB-48 | | Descriptor: | 2-[4-[[4-[1-[2-(dimethylamino)ethyl]pyrazol-4-yl]-6-[(3-methyl-1~{H}-pyrazol-5-yl)amino]pyrimidin-2-yl]amino]phenyl]-~{N}-(3-fluorophenyl)ethanamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2021-03-24 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of N-Trisubstituted Pyrimidine Derivatives as Type I RET and RET Gatekeeper Mutant Inhibitors with a Novel Kinase Binding Pose.
J.Med.Chem., 65, 2022
|
|
7ZTY
 
 | |
4YJ2
 
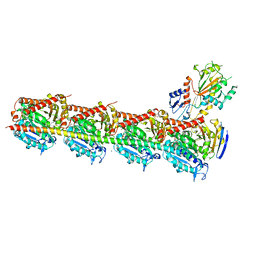 | | Crystal structure of tubulin bound to MI-181 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5,6-dimethyl-2-[(E)-2-(pyridin-3-yl)ethenyl]-1,3-benzothiazole, CALCIUM ION, ... | | Authors: | McNamara, D.E, Torres, J.Z, Yeates, T.O. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of potent anticancer compounds bound to tubulin.
Protein Sci., 24, 2015
|
|
8CUK
 
 | |
5CQ1
 
 | | Disproportionating enzyme 1 from Arabidopsis - cycloamylose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
6X8T
 
 | | CryoEM structure of the apo-SrpI encapasulin complex from Synechococcus elongatus PCC 7942 | | Descriptor: | Protein SrpI | | Authors: | LaFrance, B.J, Nichols, R.J, Phillips, N.R, Oltrogge, L.M, Valentin-Alvarado, L.E, Bischoff, A.J, Savage, D.F, Nogales, E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism.
Elife, 10, 2021
|
|
7NUV
 
 | |
5K7Q
 
 | | MicroED structure of thaumatin at 2.5 A resolution | | Descriptor: | Thaumatin-1 | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
