3TF3
 
 | |
3TH7
 
 | |
3LB9
 
 | | Crystal structure of the B. circulans cpA123 circular permutant | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | D'Angelo, I, Reitinger, S, Ludwiczek, M, Strynadka, N, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Circular permutation of Bacillus circulans xylanase: a kinetic and structural study.
Biochemistry, 49, 2010
|
|
3THJ
 
 | |
3THH
 
 | |
2ZYQ
 
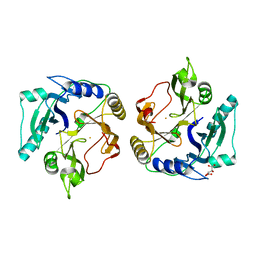 | | Crystal structure of the HsaC extradiol dioxygenase from M. tuberculosis | | Descriptor: | D(-)-TARTARIC ACID, FE (II) ION, PROBABLE BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE BPHC | | Authors: | D'Angelo, I, Yam, K.C, Eltis, L.D, Strynadka, N. | | Deposit date: | 2009-01-28 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis.
Plos Pathog., 5, 2009
|
|
3KN6
 
 | |
3KN5
 
 | |
3HJP
 
 | |
3AFE
 
 | | Crystal structure of the HsaA monooxygenase from M.tuberculosis | | Descriptor: | Hydroxylase, putative | | Authors: | D'Angelo, I, Lin, L.Y, Dresen, C, Tocheva, E.I, Eltis, L.D, Strynadka, N. | | Deposit date: | 2010-02-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism
J.Biol.Chem., 285, 2010
|
|
2VGO
 
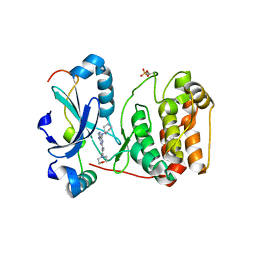 | | Crystal structure of Aurora B kinase in complex with Reversine inhibitor | | Descriptor: | INNER CENTROMERE PROTEIN A, N~6~-cyclohexyl-N~2~-(4-morpholin-4-ylphenyl)-9H-purine-2,6-diamine, SERINE/THREONINE-PROTEIN KINASE 12-A | | Authors: | D'Alise, A.M, Amabile, G, Iovino, M, Di Giorgio, F.P, Bartiromo, M, Sessa, F, Villa, F, Musacchio, A, Cortese, R. | | Deposit date: | 2007-11-15 | | Release date: | 2008-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reversine, a Novel Aurora Kinases Inhibitor, Inhibits Colony Formation of Human Acute Myeloid Leukemia Cells.
Mol.Cancer Ther., 7, 2008
|
|
2ZI8
 
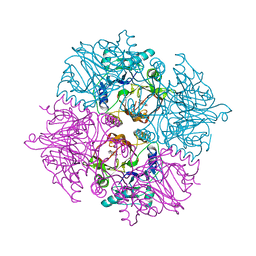 | | Crystal structure of the HsaC extradiol dioxygenase from M. tuberculosis in complex with 3,4-dihydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (DHSA) | | Descriptor: | 3,4-dihydroxy-9,10-secoandrosta-1(10),2,4-triene-9,17-dione, FE (II) ION, PROBABLE BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE BPHC | | Authors: | D'Angelo, I, Yam, K.C, Eltis, L.D, Strynadka, N. | | Deposit date: | 2008-02-13 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis.
Plos Pathog., 5, 2009
|
|
1F9A
 
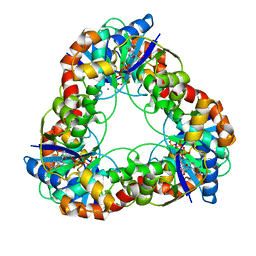 | | CRYSTAL STRUCTURE ANALYSIS OF NMN ADENYLYLTRANSFERASE FROM METHANOCOCCUS JANNASCHII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HYPOTHETICAL PROTEIN MJ0541, MAGNESIUM ION | | Authors: | D'Angelo, I, Raffaelli, N, Dabusti, V, Lorenzi, T, Magni, G, Rizzi, M. | | Deposit date: | 2000-07-09 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of nicotinamide mononucleotide adenylyltransferase: a key enzyme in NAD(+) biosynthesis.
Structure Fold.Des., 8, 2000
|
|
3AFF
 
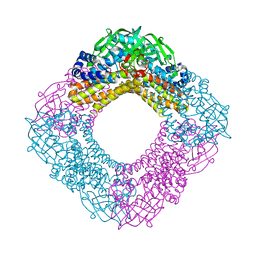 | | Crystal structure of the HsaA monooxygenase from M. tuberculosis | | Descriptor: | Hydroxylase, putative | | Authors: | D'Angelo, I, Lin, L.Y, Dresen, C, Tocheva, E.I, Strynadka, N, Eltis, L.D. | | Deposit date: | 2010-02-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism
J.Biol.Chem., 285, 2010
|
|
2ZYL
 
 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase (KshA) from M. tuberculosis | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, POSSIBLE OXIDOREDUCTASE, ... | | Authors: | D'Angelo, I, Capyk, J, Strynadka, N, Eltis, L. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of 3-ketosteroid 9{alpha}-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis
J.Biol.Chem., 284, 2009
|
|
2ESD
 
 | | Crystal Structure of thioacylenzyme intermediate of an Nadp Dependent Aldehyde Dehydrogenase | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | | Authors: | D'Ambrosio, K, Didierjean, C, Benedetti, E, Aubry, A, Corbier, C. | | Deposit date: | 2005-10-26 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The first crystal structure of a thioacylenzyme intermediate in the ALDH family: new coenzyme conformation and relevance to catalysis
Biochemistry, 45, 2006
|
|
4MNN
 
 | |
2QOA
 
 | |
2D4Q
 
 | | Crystal structure of the Sec-PH domain of the human neurofibromatosis type 1 protein | | Descriptor: | Neurofibromin, OXTOXYNOL-10, PYROPHOSPHATE 2- | | Authors: | D'angelo, I, Welti, S, Bonneau, F, Scheffzek, K. | | Deposit date: | 2005-10-22 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein
Embo Rep., 7, 2006
|
|
2QP6
 
 | |
2WVI
 
 | | Crystal Structure of the N-terminal Domain of BubR1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MITOTIC CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1 BETA, ... | | Authors: | D'Arcy, S, Davies, O.R, Blundell, T.L, Bolanos-Garcia, V.M. | | Deposit date: | 2009-10-16 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining the Molecular Basis of Bubr1 Kinetochore Interactions and Anaphase-Promoting Complex/Cyclosome (Apc/C)-Cdc20 Inhibition
J.Biol.Chem., 285, 2010
|
|
1S9S
 
 | |
2HLS
 
 | |
3V5G
 
 | |
3O27
 
 | |
