3AGP
 
 | | Structure of viral polymerase form I | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2010-04-06 | | Release date: | 2010-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Assembly of Q{beta} viral RNA polymerase with host translational elongation factors EF-Tu and -Ts
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6ZKS
 
 | | Deactive complex I, open1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6Z63
 
 | | FtsE structure from Streptococus pneumoniae in complex with ADP at 1.57 A resolution (spacegroup P 21) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE | | Authors: | Alcorlo, M, Straume, D, Havarstein, L.S, Hermoso, J.A. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae.
Mbio, 11, 2020
|
|
6Z67
 
 | | FtsE structure of Streptococcus pneumoniae in complex with AMPPNP at 2.4 A resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Alcorlo, M, Straume, D, Havarstein, L.S, Hermoso, j.A. | | Deposit date: | 2020-05-28 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae.
Mbio, 11, 2020
|
|
5JKI
 
 | | Crystal structure of the first transmembrane PAP2 type phosphatidylglycerolphosphate phosphatase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative lipid phosphate phosphatase YodM, TUNGSTATE(VI)ION, ... | | Authors: | El Ghachi, M, Howe, N, Lampion, A, Delbrassine, F, Vogeley, L, Caffrey, M, Sauvage, E, Auger, R, Guiseppe, A, Roure, S, Perlier, S, Mengin-lecreulx, D, Foglino, M, Touze, T. | | Deposit date: | 2016-04-26 | | Release date: | 2017-02-22 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and biochemical characterization of the transmembrane PAP2 type phosphatidylglycerol phosphate phosphatase from Bacillus subtilis.
Cell. Mol. Life Sci., 74, 2017
|
|
1CR3
 
 | | SOLUTION CONFORMATION OF THE (+)TRANS-ANTI-BENZO[G]CHRYSENE-DA ADDUCT OPPOSITE DT IN A DNA DUPLEX | | Descriptor: | BENZO[G]CHRYSENE, DNA (5'-D(*CP*TP*CP*TP*CP*AP*CP*TP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*GP*TP*GP*AP*GP*AP*G)-3') | | Authors: | Suri, A.K, Mao, B, Amin, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1999-08-12 | | Release date: | 2000-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of the (+)-trans-anti-benzo[g]chrysene-dA adduct opposite dT in a DNA duplex.
J.Mol.Biol., 292, 1999
|
|
6ZJ3
 
 | | Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis | | Descriptor: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Matzov, D, Halfon, H, Zimmerman, E, Rozenberg, H, Bashan, A, Gray, M.W, Yonath, A.E, Shalev-Benami, M. | | Deposit date: | 2020-06-27 | | Release date: | 2020-10-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis.
Nucleic Acids Res., 48, 2020
|
|
5J2U
 
 | | Tubulin-MMAF complex | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Waight, A.B, Bargsten, K, Doronina, S, Steinmetz, M.O, Sussman, D, Prota, A.E. | | Deposit date: | 2016-03-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Microtubule Destabilization by Potent Auristatin Anti-Mitotics.
Plos One, 11, 2016
|
|
4URP
 
 | | The Crystal structure of Nitroreductase from Saccharomyces cerevisiae | | Descriptor: | FATTY ACID REPRESSION MUTANT PROTEIN 2 | | Authors: | Song, H.-N, Woo, E.-J, Bang, S.-Y, Jung, D.-G, Park, S.-G. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Crystal Structure of the Fungal Nitroreductase Frm2 from Saccharomyces Cerevisiae.
Protein Sci., 24, 2015
|
|
3AGQ
 
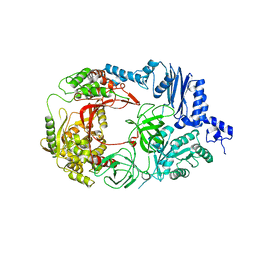 | | Structure of viral polymerase form II | | Descriptor: | Elongation factor Ts, Elongation factor Tu 1, LINKER, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2010-04-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Assembly of Q{beta} viral RNA polymerase with host translational elongation factors EF-Tu and -Ts
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5XH3
 
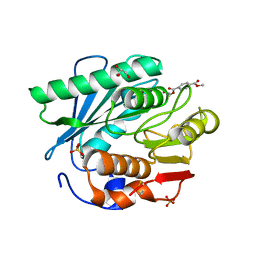 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant in complex with HEMT from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, O 4-(2-hydroxyethyl) O 1-methyl benzene-1,4-dicarboxylate, Poly(ethylene terephthalate) hydrolase, ... | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
2P61
 
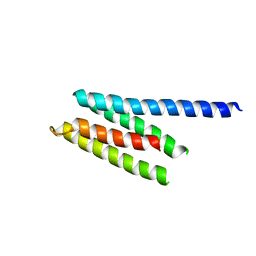 | | Crystal structure of protein TM1646 from Thermotoga maritima, Pfam DUF327 | | Descriptor: | Hypothetical protein TM_1646 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical protein TM_1646 from Thermotoga maritima
To be Published
|
|
2DVH
 
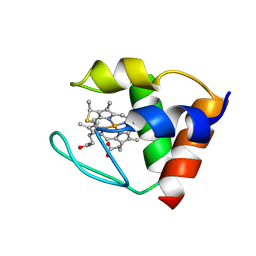 | | THE Y64A MUTANT OF CYTOCHROME C553 FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH, NMR, 39 STRUCTURES | | Descriptor: | CYTOCHROME C-553, HEME C | | Authors: | Sebban-Kreuzer, C, Blackledge, M.J, Dolla, A, Marion, D, Guerlesquin, F. | | Deposit date: | 1998-03-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of ferrocytochrome c553 from Desulfovibrio vulgaris studied by NMR spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 245, 1995
|
|
2OUP
 
 | | crystal structure of PDE10A | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ONT
 
 | | A swapped dimer of the HIV-1 capsid C-terminal domain | | Descriptor: | Capsid protein p24 | | Authors: | Ivanov, D, Tsodikov, O.V, Kasanov, J, Ellenberger, T, Wagner, G, Collins, T. | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Domain-swapped dimerization of the HIV-1 capsid C-terminal domain
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PER
 
 | |
2P5J
 
 | | sPLA2 inhibitor pip 17 | | Descriptor: | pip17 | | Authors: | Thwin, M.M, Satyanarayanajois, D.S, Nagarajarao, L.M, Sato, K, Gopalakrishnakone, P.P, Arjunan, P. | | Deposit date: | 2007-03-15 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Novel Peptide Inhibitors of Human Secretory Phospholipase A2 with Antiinflammatory Activity: Solution Structure and Molecular Modeling.
J.Med.Chem., 50, 2007
|
|
2OUR
 
 | | crystal structure of PDE10A2 mutant D674A in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUN
 
 | | crystal structure of PDE10A2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H, Ke, H.M. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2P1W
 
 | |
5XHY
 
 | | BRD4 bound with compound Bdi1 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-(4-cyclopropyl-1,3,3-trimethyl-2-oxidanylidene-quinoxalin-6-yl)-4-methyl-benzenesulfonamide | | Authors: | Xiong, B, Cao, D, Li, Y. | | Deposit date: | 2017-04-25 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | BRD4 bound with compound Bdi1
To Be Published
|
|
2OUU
 
 | | crystal structure of PDE10A2 mutant D674A in complex with cGMP | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3O6O
 
 | | Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of an the inhibitor BIIB021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein 83, PENTAETHYLENE GLYCOL | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Weadge, J, Li, Y, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P, Fairlamb, A.H, Ferguson, M.A.J, Thompson, S, MacKenzie, C, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
2DEN
 
 | |
2DSM
 
 | | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450 | | Descriptor: | Hypothetical protein yqaI | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-01 | | Release date: | 2006-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450
to be published
|
|
