4FKM
 
 | |
8SF2
 
 | | Crystal structure of the engineered SsoPox variant IG7 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
7S80
 
 | | Crystal structure of iAChSnFR Acetylcholine Sensor precursor binding protein | | Descriptor: | 1,2-ETHANEDIOL, MALONATE ION, iAchSnFR Fluorescent Acetylcholine Sensor precursor binding protein | | Authors: | Fan, C, Borden, P.M, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
8SF9
 
 | | Crystal structure of the engineered SsoPox variant IG7 - Alternative state | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
4FEP
 
 | | Crystal structure of the A24U/U25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, A24U/U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, COBALT HEXAMMINE(III) | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
2GI4
 
 | |
2G6B
 
 | | Crystal structure of human RAB26 in complex with a GTP analogue | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-26, ... | | Authors: | Wang, J, Tempel, W, Shen, Y, Shen, L, Yaniw, D, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-24 | | Release date: | 2006-03-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human RAB26 in complex with a GTP analogue
To be Published
|
|
6LD7
 
 | |
4FQ5
 
 | | Crystal structure of the maleate isomerase Iso(C200A) from Pseudomonas putida S16 with maleate | | Descriptor: | MALEIC ACID, Maleate cis-trans isomerase | | Authors: | Lu, Y, Chen, D, Zhang, Z, Li, Q, Wu, G, Xu, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside.
Mol.Microbiol., 87, 2013
|
|
8SFD
 
 | | Crystal structure of the engineered SsoPox variant IVB10 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
4WIB
 
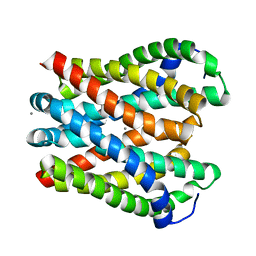 | | Crystal structure of Magnesium transporter MgtE | | Descriptor: | CALCIUM ION, Magnesium transporter MgtE | | Authors: | Takeda, H, Hattori, M, Nishizawa, T, Yamashita, K, Shah, S.T.A, Caffrey, M, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-09-25 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Nat Commun, 5, 2014
|
|
4WIH
 
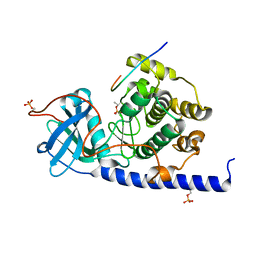 | | Crystal structure of cAMP-dependent Protein Kinase A from Cricetulus griseus | | Descriptor: | cAMP Dependent Protein Kinase Inhibitor PKI-tide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Kudlinzki, D, Linhard, V.L, Saxena, K, Dreyer, M, Schwalbe, H. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.139 Å) | | Cite: | High-resolution crystal structure of cAMP-dependent protein kinase from Cricetulus griseus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4WL3
 
 | |
4WIT
 
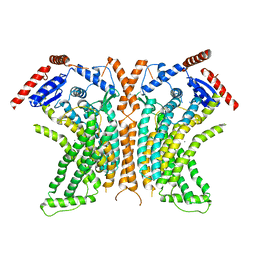 | | TMEM16 lipid scramblase in crystal form 2 | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Dutzler, R, Brunner, J.D, Lim, N.K, Schenck, S. | | Deposit date: | 2014-09-26 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | X-ray structure of a calcium-activated TMEM16 lipid scramblase.
Nature, 516, 2014
|
|
4WLJ
 
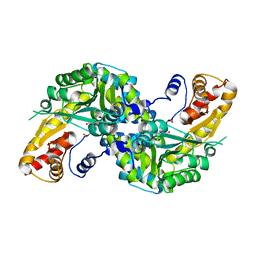 | | High resolution crystal structure of human kynurenine aminotransferase-I in complex with aminooxyacetate | | Descriptor: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | High resolution crystal structures of human kynurenine aminotransferase-I bound to PLP cofactor, and in complex with aminooxyacetate.
Protein Sci., 26, 2017
|
|
7SEY
 
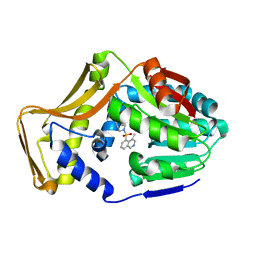 | | M. tb EgtD in complex with SGH | | Descriptor: | 2-amino-1-[(3S)-3-methyl-4-(4-methylisoquinoline-5-sulfonyl)-1,4-diazepan-1-yl]ethan-1-one, Histidine N-alpha-methyltransferase | | Authors: | Sudasinghe, T.D, Ronning, D.R. | | Deposit date: | 2021-10-02 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Inhibitors of Mycobacterium tuberculosis EgtD target both substrate binding sites to limit hercynine production.
Sci Rep, 11, 2021
|
|
4WNT
 
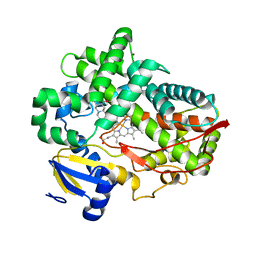 | | Human Cytochrome P450 2D6 Ajmalicine Complex | | Descriptor: | Ajmalicine, Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
4WPZ
 
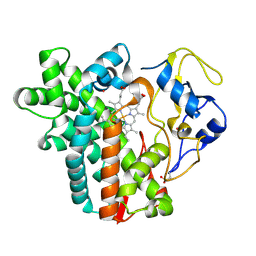 | | Crystal structure of cytochrome P450 CYP107W1 from Streptomyces avermitilis | | Descriptor: | Cytochrome P450, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kang, L.W, Kim, D.H, Pham, T.V, Han, S.H. | | Deposit date: | 2014-10-21 | | Release date: | 2015-04-22 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional characterization of CYP107W1 from Streptomyces avermitilis and biosynthesis of macrolide oligomycin A.
Arch.Biochem.Biophys., 575, 2015
|
|
7CZ5
 
 | | Cryo-EM structure of the human growth hormone-releasing hormone receptor-Gs protein complex | | Descriptor: | CHOLESTEROL, Growth hormone-releasing hormone receptor,growth hormone-releasing hormone receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhou, F, Zhang, H, Cong, Z, Zhao, L, Zhou, Q, Mao, C, Cheng, X, Shen, D, Cai, X, Ma, C, Wang, Y, Dai, A, Zhou, Y, Sun, W, Zhao, F, Zhao, S, Jiang, H, Jiang, Y, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-07 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for activation of the growth hormone-releasing hormone receptor.
Nat Commun, 11, 2020
|
|
6LDM
 
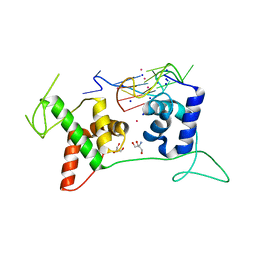 | | Structural basis of G-quadruplex DNA recognition by the yeast telomeric protein Rap1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA-binding protein RAP1, G-guadruplex DNA, ... | | Authors: | Traczyk, A, Gill, D.J, Chong, W.L, Rhodes, D. | | Deposit date: | 2019-11-22 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of G-quadruplex DNA recognition by the yeast telomeric protein Rap1.
Nucleic Acids Res., 48, 2020
|
|
7D68
 
 | | Cryo-EM structure of the human glucagon-like peptide-2 receptor-Gs protein complex | | Descriptor: | Glucagon-like peptide 2 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, W, Chen, L, Zhou, Q, Zhao, L, Zhang, H, Cong, Z, Shen, D, Zhao, F, Zhou, F, Cai, X, Chen, Y, Zhou, Y, Gadgaard, S, van der Velden, W.J, Zhao, S, Jiang, Y, Rosenkilde, M.M, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor.
Cell Res., 30, 2020
|
|
4WPG
 
 | | Group A Streptococcus GacA is an essential dTDP-4-dehydrorhamnose reductase (RmlD) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, ... | | Authors: | van der Beek, S.L, Le Breton, Y, Ferenbach, A.T, van Aalten, D.M.F, Navratilova, I, McIver, K, van Sorge, N.M, Dorfmueller, H.C. | | Deposit date: | 2014-10-18 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD).
Mol.Microbiol., 98, 2015
|
|
7S9Y
 
 | | Helicobacter Hepaticus CcsBA Open Conformation | | Descriptor: | Cytochrome c biogenesis protein, HEME B/C, PHOSPHATIDYLETHANOLAMINE | | Authors: | Mendez, D.L, Lowder, E.P, Tillman, D.E, Sutherland, M.C, Collier, A.L, Rau, M.J, Fitzpatrick, J.A, Kranz, R.G. | | Deposit date: | 2021-09-21 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM of CcsBA reveals the basis for cytochrome c biogenesis and heme transport.
Nat.Chem.Biol., 18, 2022
|
|
7DFP
 
 | | Human dopamine D2 receptor in complex with spiperone | | Descriptor: | 8-[4-(4-fluorophenyl)-4-oxidanylidene-butyl]-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, D(2) dopamine receptor,Soluble cytochrome b562, FabH, ... | | Authors: | Im, D, Shimamura, T, Iwata, S. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the dopamine D 2 receptor in complex with the antipsychotic drug spiperone.
Nat Commun, 11, 2020
|
|
7SEF
 
 | | Structure of E. coli LetB delta (Ring6) mutant, Ring 1 in the open state (Model 2, Rings 1-3 only) | | Descriptor: | MCE family protein, Intermembrane transport protein YebT chimera | | Authors: | Vieni, C, Coudray, N, Bhabha, G, Ekiert, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Role of Ring6 in the Function of the E. coli MCE Protein LetB.
J.Mol.Biol., 434, 2022
|
|
