4W6L
 
 | |
2VLG
 
 | | KinA PAS-A domain, homodimer | | 分子名称: | ACETATE ION, CHLORIDE ION, SPORULATION KINASE A | | 著者 | Lee, J, Tomchick, D.R, Brautigam, C.A, Machius, M, Kort, R, Hellingwerf, K.J, Gardner, K.H. | | 登録日 | 2008-01-14 | | 公開日 | 2008-03-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Changes at the Kina Pas-A Dimerization Interface Influence Histidine Kinase Function.
Biochemistry, 47, 2008
|
|
4W6C
 
 | |
7AFN
 
 | | Bacterial 30S ribosomal subunit assembly complex state B (head domain) | | 分子名称: | 16SrRNA (head domain of the 30S ribosome), 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | 著者 | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | 登録日 | 2020-09-19 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
4W6K
 
 | |
7AFD
 
 | | Bacterial 30S ribosomal subunit assembly complex state A (head domain) | | 分子名称: | 16SrRNA of the head domain (residue C931 to G1386), 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | 著者 | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | 登録日 | 2020-09-19 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
1K0O
 
 | | Crystal structure of a soluble form of CLIC1. An intracellular chloride ion channel | | 分子名称: | CHLORIDE INTRACELLULAR CHANNEL PROTEIN 1 | | 著者 | Harrop, S.J, DeMaere, M.Z, Fairlie, W.D, Reztsova, T, Valenzuela, S.M, Mazzanti, M, Tonini, R, Qiu, M.R, Jankova, L, Warton, K, Bauskin, A.R, Wu, W.M, Pankhurst, S, Campbell, T.J, Breit, S.N, Curmi, P.M.G. | | 登録日 | 2001-09-19 | | 公開日 | 2001-12-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution.
J.Biol.Chem., 276, 2001
|
|
4RCD
 
 | | Crystal structure of BACE1 in complex with a 2-aminooxazoline 4-azaxanthene inhibitor | | 分子名称: | (5S)-7-(2-fluoropyridin-3-yl)-3-[(3-methyloxetan-3-yl)ethynyl]spiro[chromeno[2,3-b]pyridine-5,4'-[1,3]oxazol]-2'-amine, Beta-secretase 1, GLYCEROL, ... | | 著者 | Whittington, D.A, Long, A.M. | | 登録日 | 2014-09-15 | | 公開日 | 2015-01-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Inhibitors of beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE1): Identification of (S)-7-(2-Fluoropyridin-3-yl)-3-((3-methyloxetan-3-yl)ethynyl)-5'H-spiro[chromeno[2,3-b]pyridine-5,4'-oxazol]-2'-amine (AMG-8718).
J.Med.Chem., 57, 2014
|
|
7SJX
 
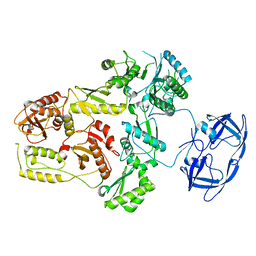 | | Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein | | 分子名称: | Gag-Pol polyprotein | | 著者 | Lyumkis, D, Passos, D, Arnold, E, Harrison, J.J.E.K, Ruiz, F.X. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | Cryo-EM structure of the HIV-1 Pol polyprotein provides insights into virion maturation.
Sci Adv, 8, 2022
|
|
7AFA
 
 | | Bacterial 30S ribosomal subunit assembly complex state F (head domain) | | 分子名称: | 16SrRNA (head domain of the 30S ribosome), 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | 著者 | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Kaminishi, T, Diercks, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | 登録日 | 2020-09-19 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
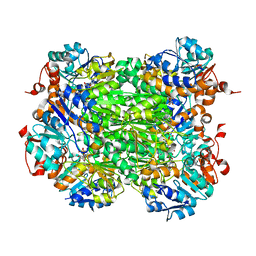
2VHW
 
 | |
6P7X
 
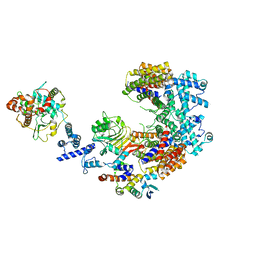 | | Structure of the K. lactis CBF3 core - Ndc10 D1D2 complex | | 分子名称: | Cep3, Ctf13, Ndc10, ... | | 著者 | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | 登録日 | 2019-06-06 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
7AFO
 
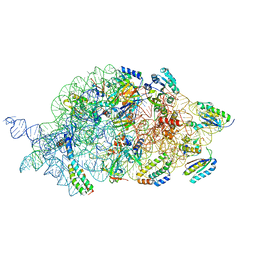 | | Bacterial 30S ribosomal subunit assembly complex state B (body domain) | | 分子名称: | 16SrRNA (body domain of the 30S ribosome), 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | 著者 | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | 登録日 | 2020-09-19 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
5JX0
 
 | |
7AFH
 
 | | Bacterial 30S ribosomal subunit assembly complex state C (head domain) | | 分子名称: | 16SrRNA (head domain of the 30S ribosome), 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | 著者 | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | 登録日 | 2020-09-19 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | 分子名称: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
5K4D
 
 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 3 | | 分子名称: | Eukaryotic translation initiation factor 3 subunit D | | 著者 | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | 登録日 | 2016-05-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
6W7V
 
 | | Structure of rabbit actin in complex with truncated analog of Mycalolide B | | 分子名称: | (1E,3R,4R,5S,6R,9S,10S,12S)-12-[(4-aminobutanoyl)oxy]-1-[ethyl(formyl)amino]-4,10-dimethoxy-3,5,9,13-tetramethyltetradec-1-en-6-yl (2R)-oxolane-2-carboxylate, 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Allingham, J.S, Deng, X, Trofimova, D. | | 登録日 | 2020-03-19 | | 公開日 | 2020-10-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Truncated Actin-Targeting Macrolide Derivative Blocks Cancer Cell Motility and Invasion of Extracellular Matrix.
J.Am.Chem.Soc., 143, 2021
|
|
2W5W
 
 | | Structure of TAB5 alkaline phosphatase mutant His 135 Asp with Zn bound in the M3 site. | | 分子名称: | ALKALINE PHOSPHATASE, ZINC ION | | 著者 | Koutsioulis, D, Lyskowski, A, Maki, S, Guthrie, E, Feller, G, Bouriotis, V, Heikinheimo, P. | | 登録日 | 2008-12-15 | | 公開日 | 2009-11-24 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Coordination Sphere of the Third Metal Site is Essential to the Activity and Metal Selectivity of Alkaline Phosphatases.
Protein Sci., 19, 2010
|
|
1JJE
 
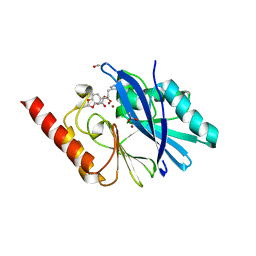 | |
7ZZ6
 
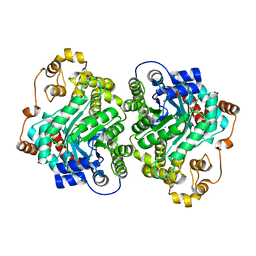 | | Cryo-EM structure of "CT-CT dimer" of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | 分子名称: | MAGNESIUM ION, MANGANESE (II) ION, PYRUVIC ACID, ... | | 著者 | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | 登録日 | 2022-05-25 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.15 Å) | | 主引用文献 | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
8IHR
 
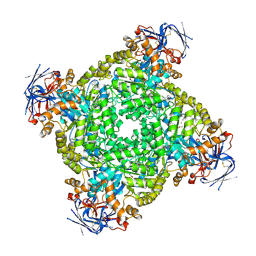 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | 分子名称: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
1JK0
 
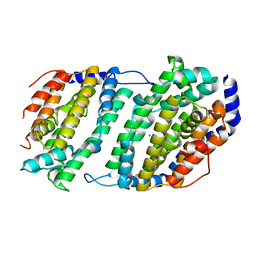 | | Ribonucleotide reductase Y2Y4 heterodimer | | 分子名称: | ZINC ION, ribonucleoside-diphosphate reductase small chain 1, ribonucleoside-diphosphate reductase small chain 2 | | 著者 | Voegtli, W.C, Perlstein, D.L, Ge, J, Stubbe, J, Rosenzweig, A.C. | | 登録日 | 2001-07-10 | | 公開日 | 2001-09-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of the yeast ribonucleotide reductase Y2Y4 heterodimer.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5DDJ
 
 | | Crystal structure of recombinant foot-and-mouth-disease virus O1M-S2093Y empty capsid | | 分子名称: | Foot and mouth disease virus, VP1, VP2, ... | | 著者 | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | 登録日 | 2015-08-25 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure-based energetics of protein interfaces guides foot-and-mouth disease virus vaccine design.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | 分子名称: | Amidohydrolase family protein, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
