6YCM
 
 | | Crystal structure of GcoA T296S bound to p-vanillin | | 分子名称: | 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, cytochrome P450 subunit, ... | | 著者 | Mallinson, S.J.B, Hinchen, D.J, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | 登録日 | 2020-03-18 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
7Q0O
 
 | | E. coli NfsA | | 分子名称: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NADPH nitroreductase | | 著者 | White, S.A, Grainger, A, Parr, R, Day, M.A, Jarrom, D, Graziano, A, Searle, P.F, Hyde, E.I. | | 登録日 | 2021-10-15 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.96 Å) | | 主引用文献 | The 3D-structure, kinetics and dynamics of the E. coli nitroreductase NfsA with NADP + provide glimpses of its catalytic mechanism.
Febs Lett., 596, 2022
|
|
6VSA
 
 | |
7KYC
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf1-Lem3 complex in the E2P state | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | 登録日 | 2020-12-07 | | 公開日 | 2021-01-06 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
8HHM
 
 | | Cryo-EM structure of the Cas12m2-crRNA-target DNA ternary complex intermediate state | | 分子名称: | Cas12m2, DNA (36-MER), MAGNESIUM ION, ... | | 著者 | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | 登録日 | 2022-11-16 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BO6
 
 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 2 | | 分子名称: | (~{E})-~{N}-[[5-(3-azanyl-1~{H}-indazol-6-yl)-4-chloranyl-1~{H}-imidazol-2-yl]methyl]-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enamide, CITRIC ACID, Coagulation factor XIa light chain, ... | | 著者 | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | 登録日 | 2022-11-14 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
5K75
 
 | | IRAK4 in complex with Compound 1 | | 分子名称: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}1-(7,8-dihydro-6~{H}-cyclopenta[2,3]thieno[2,4-~{c}]pyrimidin-1-yl)-~{N}4,~{N}4-dimethyl-cyclohexane-1,4-diamine | | 著者 | Ferguson, A.D. | | 登録日 | 2016-05-25 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
7PIZ
 
 | |
8BO7
 
 | | COAGULATION FACTOR XI PROTEASE DOMAIN IN COMPLEX WITH ACTIVE SITE INHIBITOR 34 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-[[(2~{S})-2-[4-(5-chloranyl-2-cyano-phenyl)-3-methoxy-6-oxidanylidene-2,5-dihydropyridin-1-yl]-3-[(2~{S})-oxan-2-yl]propanoyl]amino]benzoic acid, CITRIC ACID, ... | | 著者 | Schaefer, M, Roehrig, S, Ackerstaff, J, Nunez, E.J, Gericke, K.M, Meier, K, Tersteegen, A, Stampfuss, J, Ellerbrock, P, Meibom, D, Lang, D, Heitmeier, S, Hillisch, A. | | 登録日 | 2022-11-14 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Design and Preclinical Characterization Program toward Asundexian (BAY 2433334), an Oral Factor XIa Inhibitor for the Prevention and Treatment of Thromboembolic Disorders.
J.Med.Chem., 66, 2023
|
|
6W3W
 
 | | An enumerative algorithm for de novo design of proteins with diverse pocket structures | | 分子名称: | DENOVO NTF2, NITRATE ION | | 著者 | Bera, A.K, Basanta, B, Dimaio, F, Sankaran, B, Baker, D. | | 登録日 | 2020-03-09 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4FG4
 
 | |
7KY5
 
 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the E2P transition state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, CHOLESTEROL, ... | | 著者 | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | 登録日 | 2020-12-07 | | 公開日 | 2021-01-06 | | 実験手法 | ELECTRON MICROSCOPY (3.98 Å) | | 主引用文献 | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
7SL5
 
 | |
1BFB
 
 | | BASIC FIBROBLAST GROWTH FACTOR COMPLEXED WITH HEPARIN TETRAMER FRAGMENT | | 分子名称: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, BASIC FIBROBLAST GROWTH FACTOR | | 著者 | Faham, S, Rees, D.C. | | 登録日 | 1995-12-12 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Heparin structure and interactions with basic fibroblast growth factor.
Science, 271, 1996
|
|
8B4F
 
 | | Crystal structure of human cathepsin L forming a thiohemiacetal with N-Boc-2-aminoacetaldehyde | | 分子名称: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | 登録日 | 2022-09-20 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
4FQR
 
 | |
6MKF
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the imipenem-bound form | | 分子名称: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, SULFATE ION, penicillin binding protein 5 (PBP5) | | 著者 | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | 登録日 | 2018-09-25 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
7YM0
 
 | | Lysoplasmalogen-specific phospholipase D (LyPls-PLD) with Ca2+ | | 分子名称: | CALCIUM ION, Lysoplasmalogenase | | 著者 | Yasutake, Y, Sakasegawa, S, Sugimori, D, Murayama, K. | | 登録日 | 2022-07-27 | | 公開日 | 2023-01-04 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Structural basis for the substrate specificity switching of lysoplasmalogen-specific phospholipase D from Thermocrispum sp. RD004668.
Biosci.Biotechnol.Biochem., 87, 2022
|
|
8B30
 
 | |
5N5F
 
 | |
7SKZ
 
 | |
7KMZ
 
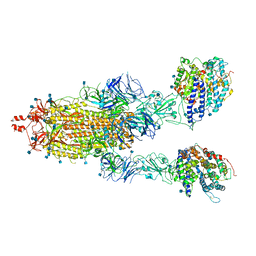 | | Cryo-EM structure of double ACE2-bound SARS-CoV-2 trimer Spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-03 | | 公開日 | 2020-12-09 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6W5H
 
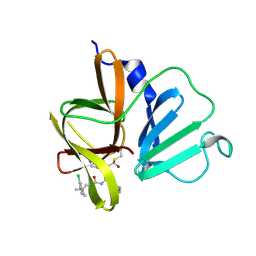 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5d | | 分子名称: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-03-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6MH2
 
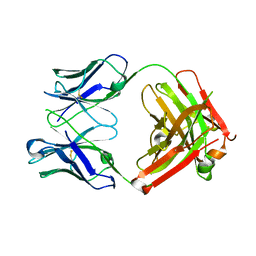 | | Structure of Herceptin Fab without antigen | | 分子名称: | Herceptin Fab arm heavy chain, Herceptin Fab arm light chain | | 著者 | Luthra, A, Langley, D.B, Christie, M, Christ, D. | | 登録日 | 2018-09-17 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Human Antibody Bispecifics through Phage Display Selection.
Biochemistry, 58, 2019
|
|
6SP6
 
 | | Ultra-high Resolution Crystal Structure of the CTX-M-15 Extended-Spectrum beta-Lactamase in Complex with Taniborbactam (VNRX-5133) | | 分子名称: | (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | 登録日 | 2019-08-31 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Discovery of Taniborbactam (VNRX-5133): A Broad-Spectrum Serine- and Metallo-beta-lactamase Inhibitor for Carbapenem-Resistant Bacterial Infections.
J.Med.Chem., 63, 2020
|
|
