5NJQ
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 4ET8 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | 登録日 | 2017-03-29 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5NJP
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 1HEW | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | 登録日 | 2017-03-29 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
7USB
 
 | | CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2022-04-23 | | 公開日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7US9
 
 | | CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2022-04-23 | | 公開日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
6QMT
 
 | | Complement factor D in complex with the inhibitor 2-(2-(3'-(aminomethyl)-[1,1'-biphenyl]-3-carboxamido)phenyl)acetic acid | | 分子名称: | 2-[2-[[3-[3-(aminomethyl)phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, Complement factor D | | 著者 | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | 登録日 | 2019-02-08 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
5NRE
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - a3GalT in complex with lactose - a3GalT-LAT | | 分子名称: | N-acetyllactosaminide alpha-1,3-galactosyltransferase, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | 登録日 | 2017-04-22 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7US5
 
 | | X-ray crystal structure of GDP-D-glycero-D-manno-heptose 4,6-Dehydratase from Campylobacter jejuni | | 分子名称: | 1,2-ETHANEDIOL, GDP-D-GLYCERO-D-MANNO-HEPTOSE 4,6-DEHYDRATASE, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Thoden, J.B, Xiang, D.F, Raushel, F.M, Holden, H.M. | | 登録日 | 2022-04-23 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Reaction Mechanism and Three-Dimensional Structure of GDP-d-glycero-alpha-d-manno-heptose 4,6-Dehydratase from Campylobacter jejuni.
Biochemistry, 61, 2022
|
|
6QP9
 
 | | Drosophila Semaphorin 1a, extracellular domains 1-2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin-1A | | 著者 | Rozbesky, D, Robinson, R.A, Harlos, K, Siebold, C, Jones, E.Y. | | 登録日 | 2019-02-13 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
6W1R
 
 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.23 Angstrom resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2020-03-04 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NRD
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - alpha-3GalT in complex with Co2+, UDP-Gal and lactose - a3GalT-Co2+-UDP-Gal-LAT-2 | | 分子名称: | COBALT (II) ION, GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | 登録日 | 2017-04-22 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7V19
 
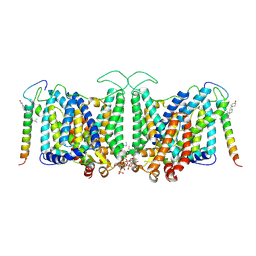 | | Local refinement of Band 3-II transmembrane domains, class 1 of erythrocyte ankyrin-1 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHOLESTEROL, ... | | 著者 | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | 登録日 | 2022-05-11 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-07-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6OJP
 
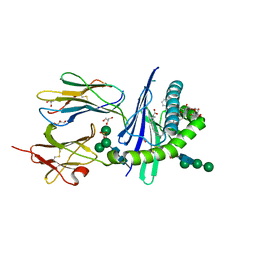 | | Structure of glycolipid alpha-GSA[8,6P] in complex with mouse CD1d | | 分子名称: | (5R,6S,7S)-5,6-dihydroxy-7-(octanoylamino)-N-(6-phenylhexyl)-8-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}octanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | 著者 | Zajonc, D.M, Bitra, A. | | 登録日 | 2019-04-11 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6WPS
 
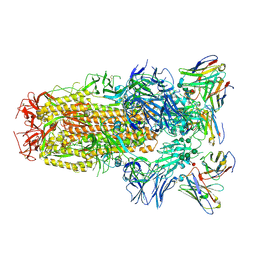 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | 著者 | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-04-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
8VTX
 
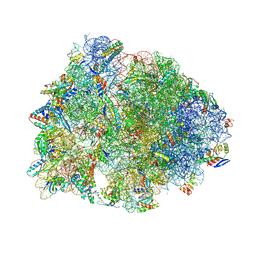 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with macrolone MCX-128, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.40A resolution | | 分子名称: | 1-cyclopropyl-7-(4-{4-[({[(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-14-(methoxyimino)-3a,7,9,11,13,15-hexamethyl-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl]oxy}carbonyl)amino]butyl}piperazin-1-yl)-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | 著者 | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | 登録日 | 2024-01-27 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 2024
|
|
8VTY
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with ciprofloxacin and protein Y at 2.60A resolution | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, 16S Ribosomal RNA, ... | | 著者 | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | 登録日 | 2024-01-27 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 2024
|
|
8WJN
 
 | | Cryo-EM structure of 6-subunit Smc5/6 head region | | 分子名称: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosomes element 1, Non-structural maintenance of chromosomes element 4, ... | | 著者 | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | 登録日 | 2023-09-26 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (5.58 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8W2C
 
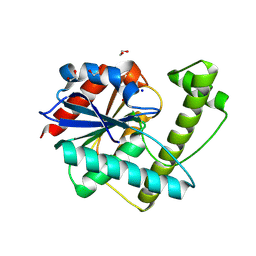 | |
8XGK
 
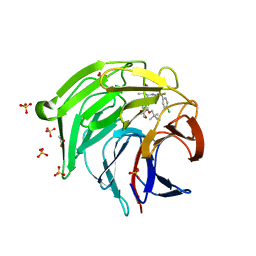 | | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Ihhibitors | | 分子名称: | (2~{R},3~{S})-3-[[(2~{S})-2-(4-chlorophenyl)-2-fluoranyl-ethanoyl]amino]-3-[3-(2-cyano-2-methyl-propoxy)-4-methyl-phenyl]-2-methyl-propanoic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | 著者 | Otake, K, Hara, Y, Ubukata, M, Inoue, M, Nagahashi, N, Motoda, D, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | 登録日 | 2023-12-15 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Inhibitors.
J.Med.Chem., 67, 2024
|
|
8X4F
 
 | |
9FE7
 
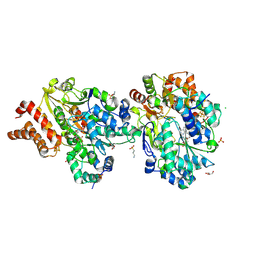 | | Crystal Structure of oxidized NuoEF variant P228R(NuoF) from Aquifex aeolicus | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | 登録日 | 2024-05-17 | | 公開日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
8ZFO
 
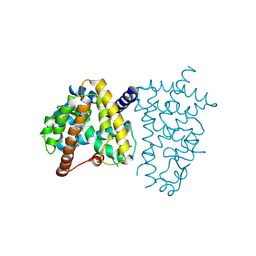 | |
9EUN
 
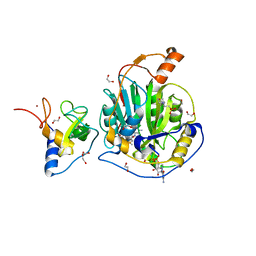 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with SAM and m7GTP | | 分子名称: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | 登録日 | 2024-03-27 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9BUL
 
 | |
8ZFR
 
 | |
9AZX
 
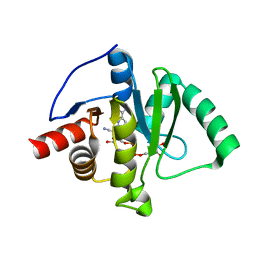 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with NDPr | | 分子名称: | Non-structural protein 3, {(2R,3S,4R,5R)-5-[(8S)-4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl]-5-cyano-3,4-dihydroxyoxolan-2-yl}methyl [(2R,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methyl dihydrogen diphosphate | | 著者 | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | 登録日 | 2024-03-11 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.395 Å) | | 主引用文献 | GS-441524-Diphosphate-Ribose Derivatives as Nanomolar Binders and Fluorescence Polarization Tracers for SARS-CoV-2 and Other Viral Macrodomains.
Acs Chem.Biol., 19, 2024
|
|
