6BOJ
 
 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with BPN5004 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-{[2-(3-chlorophenyl)-6-ethylpyrimidin-4-yl]methyl}phenyl)acetamide, CHLORIDE ION, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2017-11-20 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Memory enhancing effects of BPN14770, an allosteric inhibitor of phosphodiesterase-4D, in wild-type and humanized mice.
Neuropsychopharmacology, 43, 2018
|
|
4TZN
 
 | | Structure of HTP-2 bound to HTP-3 motif-6 | | Descriptor: | Protein HTP-2, Protein HTP-3 | | Authors: | Rosenberg, S.C, Corbett, K.D. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.115 Å) | | Cite: | The Chromosome Axis Controls Meiotic Events through a Hierarchical Assembly of HORMA Domain Proteins.
Dev.Cell, 31, 2014
|
|
7LPR
 
 | |
5O4Z
 
 | | Structure of the inactive T.maritima PDE (TM1595) D80N D154N mutant with substrate 5'-pApA | | Descriptor: | ADENOSINE-5'-MONOPHOSPHATE, CHLORIDE ION, DHH/DHHA1-type phosphodiesterase TM1595, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-05-31 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
5HS0
 
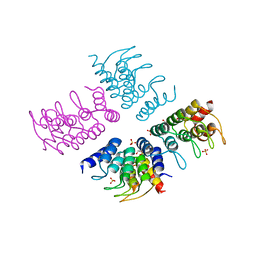 | | Computationally Designed Cyclic Tetramer ank1C4_7 | | Descriptor: | Ankyrin domain protein ank1C4_7, GLYCEROL, SULFATE ION | | Authors: | McNamara, D.E, Cascio, D, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5DTD
 
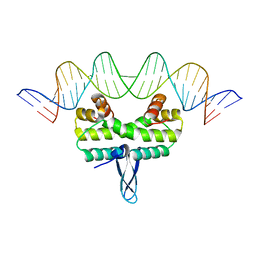 | |
1A4Z
 
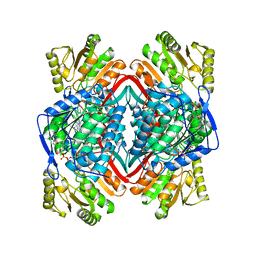 | |
5J8D
 
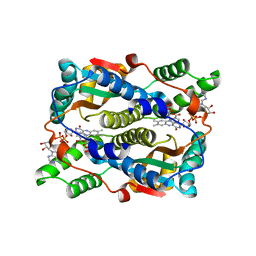 | | Structure of nitroreductase from E. cloacae complexed with nicotinic acid adenine dinucleotide | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID ADENINE DINUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2016-04-07 | | Release date: | 2017-05-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism-Informed Refinement Reveals Altered Substrate-Binding Mode for Catalytically Competent Nitroreductase.
Structure, 25, 2017
|
|
6QSD
 
 | | Crystal structure of Pizza6S | | Descriptor: | Pizza6S, SULFATE ION | | Authors: | Noguchi, H, Vandebroek, L, Kamata, K, Tame, J.R.H, Van Meervelt, L, Parac-Vogt, T.N, Voet, A.R.D. | | Deposit date: | 2019-02-20 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Hybrid assemblies of a symmetric designer protein and polyoxometalates with matching symmetry.
Chem.Commun.(Camb.), 56, 2020
|
|
6QSH
 
 | | Crystal structure of the hybrid bioinorganic complex of Pizza6S linked by the 1:2 Ce-substituted Keggin | | Descriptor: | 1:2 Ce-substituted Keggin, Pizza6S | | Authors: | Noguchi, H, Vandebroek, L, Kamata, K, Tame, J.R.H, Van Meervelt, L, Parac-Vogt, T.N, Voet, A.R.D. | | Deposit date: | 2019-02-20 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Hybrid assemblies of a symmetric designer protein and polyoxometalates with matching symmetry.
Chem.Commun.(Camb.), 56, 2020
|
|
6YXO
 
 | |
4U07
 
 | | ATP bound to eukaryotic FIC domain containing protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, ... | | Authors: | Cole, A.R, Bunney, T.D, Katan, M. | | Deposit date: | 2014-07-11 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions.
Structure, 22, 2014
|
|
7UJI
 
 | |
6CP5
 
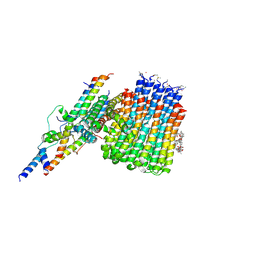 | | Monomer yeast ATP synthase Fo reconstituted in nanodisc with inhibitor of oligomycin bound generated from focused refinement. | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-11 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
5HYL
 
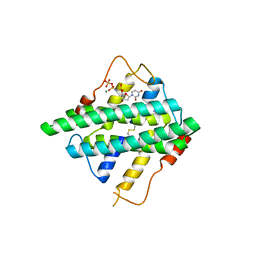 | | Crystal structure of DR2231_E47A mutant in complex with dUMPNPP and magnesium | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DR2231, MAGNESIUM ION | | Authors: | Mota, C.S, Goncalves, A.M.D, de Sanctis, D. | | Deposit date: | 2016-02-01 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deinococcus radiodurans DR2231 is a two-metal-ion mechanism hydrolase with exclusive activity on dUTP.
FEBS J., 283, 2016
|
|
1ADU
 
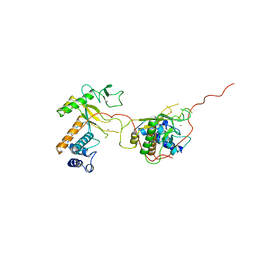 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Tucker, P.A, Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C. | | Deposit date: | 1995-05-11 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
6YXP
 
 | |
6N17
 
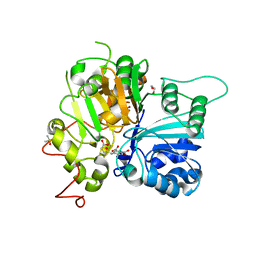 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ577 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-carboxypropanoyl)amino]benzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6ZDP
 
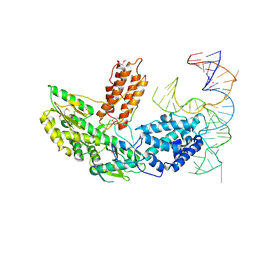 | | Structure of telomerase from Candida Tropicalis in complexe with TWJ fragment of telomeric RNA | | Descriptor: | Chains: B, POTASSIUM ION, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-15 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
8AMZ
 
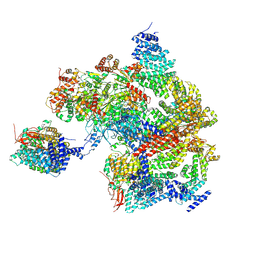 | | Spinach 19S proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1 homolog, 26S proteasome non-ATPase regulatory subunit 2 homolog, 26S proteasome regulatory subunit 7, ... | | Authors: | Kandolf, S, Grishkovskaya, I, Meinhart, A, Haselbach, D. | | Deposit date: | 2022-08-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the plant 26S proteasome
Plant Communications, 3, 2022
|
|
5NS8
 
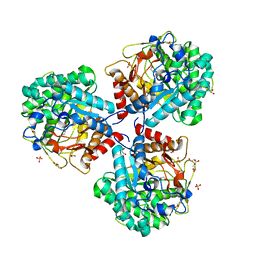 | | Crystal structure of beta-glucosidase BglM-G1 mutant H75R from marine metagenome in complex with inhibitor 1-Deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, GLYCEROL, SULFATE ION, ... | | Authors: | Mhaindarkar, D.C, Gasper, R, Lupilova, N, Leichert, L.I, Hofmann, E. | | Deposit date: | 2017-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Loss of a conserved salt bridge in bacterial glycosyl hydrolase BgIM-G1 improves substrate binding in temperate environments.
Commun Biol, 1, 2018
|
|
4TZM
 
 | |
1AMP
 
 | | CRYSTAL STRUCTURE OF AEROMONAS PROTEOLYTICA AMINOPEPTIDASE: A PROTOTYPICAL MEMBER OF THE CO-CATALYTIC ZINC ENZYME FAMILY | | Descriptor: | AMINOPEPTIDASE, ZINC ION | | Authors: | Chevrier, B, Schalk, C, D'Orchymont, H, Rondeau, J.M, Moras, D, Tarnus, C. | | Deposit date: | 1994-04-22 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Aeromonas proteolytica aminopeptidase: a prototypical member of the co-catalytic zinc enzyme family.
Structure, 2, 1994
|
|
6WX5
 
 | | Adducts formed after 3 weeks in the reaction of chlorido[chlorido(2,2'-((2-([2,2':6',2''-Terpyridin]-4'-yloxy)ethyl)azanediyl)bis(ethan-1-ol))platinum(II)] with HEWL | | Descriptor: | ACETATE ION, Lysozyme, PLATINUM (II) ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2020-05-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mustards-Derived Terpyridine-Platinum Complexes as Anticancer Agents: DNA Alkylation vs Coordination.
Inorg.Chem., 60, 2021
|
|
4YAE
 
 | | Crystal structure of LigL-apo form from Sphingobium sp. strain SYK-6 | | Descriptor: | C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
