6QDZ
 
 | | P38 alpha complex with AR117045 | | Descriptor: | 1-[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]-3-[(1~{S},4~{S})-4-[(3-propan-2-yl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)oxy]-1,2,3,4-tetrahydronaphthalen-1-yl]urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Brown, D.G, Hurley, C, Irving, S.L. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | P38 alpha complex with AR117045
To Be Published
|
|
7P6C
 
 | | Limbic-predominant neuronal inclusion body 4R tauopathy type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6B
 
 | | Limbic-predominant neuronal inclusion body 4R tauopathy type 1b tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
6QEJ
 
 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX WITH AN INHIBITOR Thiophene-2-sulfonic acid (4-fluoro-benzyl)-(4H-[1,2,4]triazol-3-ylmethyl)-amide | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Methionine aminopeptidase 2, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2019-06-05 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
8IJQ
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide | | Descriptor: | N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5FQH
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
8IH6
 
 | |
7OKW
 
 | | 1.62A X-ray crystal structure of the conserved C-terminal (CCT) of human OSR1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Bax, B.D, Elvers, K.T, Lipka-Lloyd, M, Mehellou, Y. | | Deposit date: | 2021-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of the Human SPAK and OSR1 Conserved C-Terminal (CCT) Domains.
Chembiochem, 23, 2022
|
|
6VM4
 
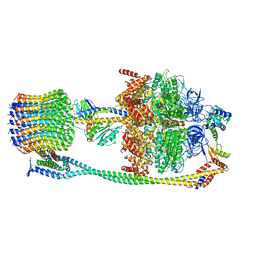 | | Chloroplast ATP synthase (C2, CF1FO) | | Descriptor: | ATP synthase delta chain, chloroplastic, ATP synthase epsilon chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.08 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6RO6
 
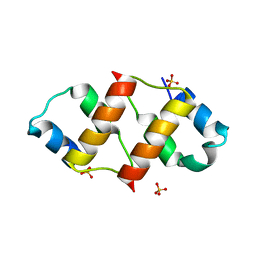 | | Crystal structure of the C-terminal dimerization domain of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | HTH-type transcriptional regulator DdrOC, SULFATE ION | | Authors: | Pignol, D, Arnoux, P, Siponen, M.I, Brandelet, G, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
6YKO
 
 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with Compound 11a | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-04-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Synthesis and structure-activity studies of novel anhydrohexitol-based Leucyl-tRNA synthetase inhibitors.
Eur.J.Med.Chem., 211, 2021
|
|
4X7F
 
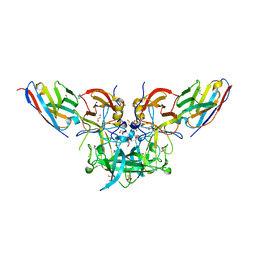 | |
6VON
 
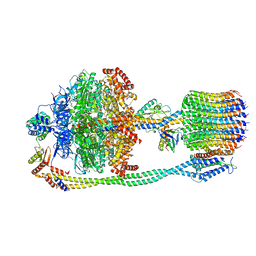 | | Chloroplast ATP synthase (R1, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
7BDW
 
 | | Crystal structure of mycobacterial PptAb in complex with ACP and compound 8918 | | Descriptor: | COENZYME A, MANGANESE (II) ION, N-(2,6-diethylphenyl)-N'-(N-ethylcarbamimidoyl)urea, ... | | Authors: | Carivenc, C, Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2020-12-22 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phosphopantetheinyl transferase binding and inhibition by amidino-urea and hydroxypyrimidinethione compounds.
Sci Rep, 11, 2021
|
|
5JLT
 
 | | The crystal structure of the bacteriophage T4 MotA C-terminal domain in complex with dsDNA reveals a novel protein-DNA recognition motif | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*TP*GP*CP*TP*TP*AP*AP*TP*AP*AP*TP*CP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*GP*AP*TP*TP*AP*TP*TP*AP*AP*GP*CP*AP*AP*AP*GP*CP*TP*TP*C)-3'), Middle transcription regulatory protein motA | | Authors: | Cuypers, M.G, Robertson, R.M, Knipling, L, Hinton, D.M, White, S.W. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The phage T4 MotA transcription factor contains a novel DNA binding motif that specifically recognizes modified DNA.
Nucleic Acids Res., 46, 2018
|
|
6QIH
 
 | |
6Q4M
 
 | | Crystal structure of the O-GlcNAc transferase Asn648Tyr mutation | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, PHOSPHATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Gundogdu, M, van Aalten, D.M.F. | | Deposit date: | 2018-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the O-GlcNAc transferase Asn648Tyr mutation
To Be Published
|
|
5A59
 
 | | The structure of GH101 E796Q mutant from Streptococcus pneumoniae TIGR4 in complex with T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
4YBI
 
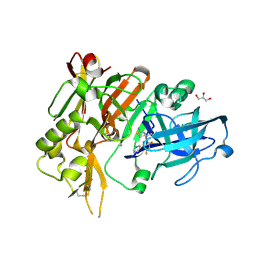 | | Crystal structure of BACE with amino thiazine inhibitor LY2811376 | | Descriptor: | (4S)-4-[2,4-difluoro-5-(pyrimidin-5-yl)phenyl]-4-methyl-5,6-dihydro-4H-1,3-thiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Timm, D.E. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Robust central reduction of amyloid-beta in humans with an orally available, non-peptidic beta-secretase inhibitor.
J.Neurosci., 31, 2011
|
|
7JWY
 
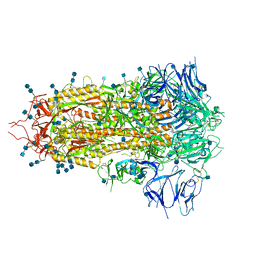 | | Structure of SARS-CoV-2 spike at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Kwong, P.D. | | Deposit date: | 2020-08-26 | | Release date: | 2020-11-25 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
8E9N
 
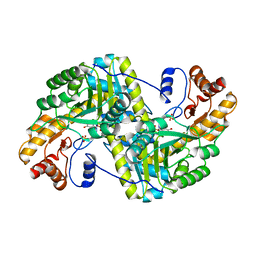 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIY in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
5AKN
 
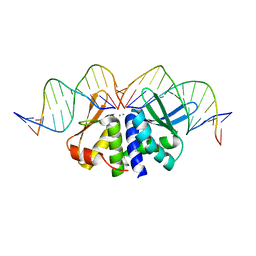 | | THE CRYSTAL STRUCTURE OF I-DMOI Q42AK120M IN COMPLEX WITH ITS TARGET DNA NICKED IN THE non-CODING STRAND B AND IN THE PRESENCE OF 2MM MN | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*CP)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP *CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
6QE1
 
 | | P38 alpha complex with AR117046 | | Descriptor: | 1-[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]-3-[(1~{S},4~{R})-4-[(3-propan-2-yl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)oxy]-1,2,3,4-tetrahydronaphthalen-1-yl]urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Brown, D.G, Hurley, C, Irving, S.L. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | P38 alpha complex with AR117045 and AR117046
To Be Published
|
|
8E9M
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT bound to maleic acid at 278 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
6QED
 
 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX; WITH AN INHIBITOR (S)-3-Hydroxy-2-oxo-1-(2-oxo-1,2,3,4-tetrahydro-quinolin-6-yl)-pyrrolidine-3-carboxylic acid 3-chloro-5-fluoro-benzylamide | | Descriptor: | (3~{S})-~{N}-[(3-chloranyl-5-fluoranyl-phenyl)methyl]-3-oxidanyl-2-oxidanylidene-1-(2-oxidanylidene-3,4-dihydro-1~{H}-quinolin-6-yl)pyrrolidine-3-carboxamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
