2MS3
 
 | |
2N30
 
 | | Structure of Ace-pvhct-NH2 | | Descriptor: | Hemocyanin subunit L2 | | Authors: | Petit, V.W, Rolland, J, Blond, A, Djediat, C, Peduzzi, J, Goulard, C, Bachere, E, Dupont, J, Destoumieux-Garzon, D, Rebuffat, S. | | Deposit date: | 2015-05-19 | | Release date: | 2015-06-17 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | A hemocyanin-derived antimicrobial peptide from the penaeid shrimp adopts an alpha-helical structure that specifically permeabilizes fungal membranes.
Biochim.Biophys.Acta, 1860, 2015
|
|
2MZP
 
 | | Structure and dynamics of the acidosis-resistant a162H mutant of the switch region of troponin I bound to the regulatory domain of troponin C | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles, ... | | Authors: | Pineda Sanabria, S.E, Robertson, I.M, Sykes, B.D. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Acidosis-Resistant A162H Mutant of the Switch Region of Troponin I Bound to the Regulatory Domain of Troponin C.
Biochemistry, 54, 2015
|
|
2MWB
 
 | | FBP28 WW2 mutant W457F | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2N2L
 
 | | NMR structure of yersinia pestis ail (attachment invasion locus) in decylphosphocholine micelles calculated with implicit membrane solvation | | Descriptor: | Outer membrane protein X | | Authors: | Marassi, F.M, Ding, Y, Tian, Y, Schwieters, C.D, Yao, Y. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone structure of Yersinia pestis Ail determined in micelles by NMR-restrained simulated annealing with implicit membrane solvation.
J.Biomol.Nmr, 63, 2015
|
|
2MOU
 
 | |
2N2R
 
 | | NMR solution structure of RsAFP2 | | Descriptor: | Defensin-like protein 2 | | Authors: | Harvey, P.J, Craik, D.J, Vriens, K. | | Deposit date: | 2015-05-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The radish defensins RsAFP1 and RsAFP2 act synergistically with caspofungin against Candida albicans biofilms.
Peptides, 75, 2016
|
|
2N2U
 
 | | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358 | | Descriptor: | OR358 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-05-14 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358
To be Published
|
|
6RJ8
 
 | | Structure of the alpha-beta hydrolase CorS from Tabernathe iboga | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Farrow, S.C, Caputi, L, Kamileen, M.O, Bussey, K, Stevenson, C.E.M, Mundy, J, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis of cycloaddition in biosynthesis of iboga and aspidosperma alkaloids.
Nat.Chem.Biol., 16, 2020
|
|
9EII
 
 | | Import stalled PINK1 TOM complex, symmetry expanded | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM20 homolog, Mitochondrial import receptor subunit TOM22 homolog, ... | | Authors: | Kirk, N.S, Glukhova, A, Callegari, S, Komander, D. | | Deposit date: | 2024-11-26 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure of human PINK1 at a mitochondrial TOM-VDAC array.
Science, 2025
|
|
6XT4
 
 | |
6LTW
 
 | | Crystal structure of Apo form of I122A/I330A variant of S-adenosylmethionine synthetase from Cryptosporidium hominis | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, S-adenosylmethionine synthase | | Authors: | Singh, R.K, Michailidou, F, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineered SAM Synthetases for Enzymatic Generation of AdoMet Analogs with Photocaging Groups and Reversible DNA Modification in Cascade Reactions.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
9GUY
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54571098 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-20 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9GUD
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54570922 | | Descriptor: | (3~{S})-3-azanyl-4-[(3~{R},4~{R},6~{S})-3-[1,3-dimethyl-2,6-bis(oxidanylidene)purin-7-yl]-4-methyl-4,6-bis(oxidanyl)azepan-1-yl]-4-oxidanylidene-butanoic acid, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-19 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9GWO
 
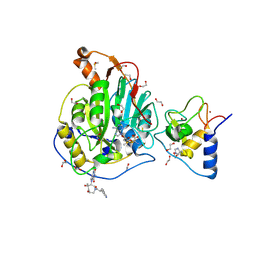 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54571126 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-27 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9FCR
 
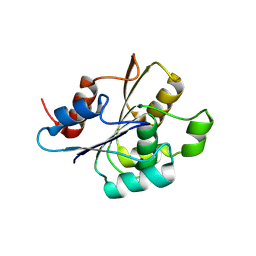 | |
9MQP
 
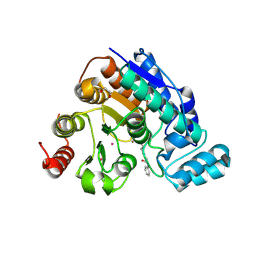 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with SelSA | | Descriptor: | 1,2-ETHANEDIOL, Hdac6 protein, N-phenyl-6-selanylhexanamide, ... | | Authors: | Goulart Stollmaier, J, Czarnecki, B.A.R, Christianson, D.W. | | Deposit date: | 2025-01-04 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanism-Based Inhibition of Histone Deacetylase 6 by a Selenocyanate Is Subject to Redox Modulation.
J.Am.Chem.Soc., 147, 2025
|
|
9FTX
 
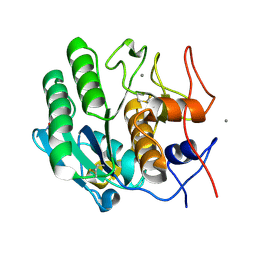 | | Serial microseconds crystallography at ID29 using fixed-target (small foils): Proteinase K with 10 um spacing between X-ray pulses | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Orlans, J, Rose, S.L, Basu, S, de Sanctis, D. | | Deposit date: | 2024-06-25 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Advancing macromolecular structure determination with microsecond X-ray pulses at a 4th generation synchrotron.
Commun Chem, 8, 2025
|
|
9F1K
 
 | | First bromodomain of BRD4 in complex with ISOX-DUAL based inhibitor 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[2-[2-[2-[4-[3-(dimethylamino)propoxy]phenyl]ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzimidazol-1-yl]ethyl]piperazin-1-yl]-N-(2-methoxyethyl)ethanamide, Bromodomain-containing protein 4 | | Authors: | Balourdas, D.I, Edmonds, A.K, Marsh, G.P, Maple, H.J, Spencer, J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | First bromodomain of BRD4 in complex with ISOX-DUAL based inhibitor 30
to be published
|
|
8A4F
 
 | | Human Interleukin-4 mutant - C3T-IL4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Mueller, T.D, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2022-06-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
6RLE
 
 | | Crystal structure of human monoamine oxidase B in complex with styrylpiperidine analogue 97 | | Descriptor: | 4-[2-(4-propan-2-ylphenyl)ethyl]-1-[(~{E})-prop-1-enyl]piperidine, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Knez, D, Colettis, N, Sova, M, Pislar, A, Higgs, J, Kamecki, F, Mangialavori, I, Dolsak, A, Zakelj, S, Trontelj, J, Kos, J, Marder, N.M, Gobec, S, Binda, C. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stereoselective Activity of 1-Propargyl-4-styrylpiperidine-like Analogues That Can Discriminate between Monoamine Oxidase Isoforms A and B.
J.Med.Chem., 63, 2020
|
|
6RLU
 
 | | Trypanosoma brucei Seryl-tRNA Synthetase in Complex with 5'-O-(N-(L-seryl)-sulfamoyl)cytidine | | Descriptor: | 5'-O-(N-(L-seryl)-sulfamoyl)cytidine, GLYCEROL, MALONATE ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
9FV0
 
 | | MsbA in MSP1D1 Nanodisc inward-facing narrow open | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shearer, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 2025
|
|
9FRL
 
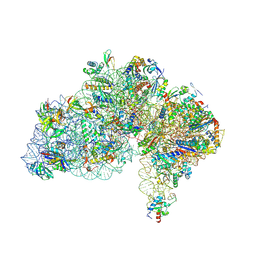 | | Cryo-EM structure of Saccharolobus solfataricus 30S initiation complex bound to SD mRNA with h44 in up position | | Descriptor: | LSU ribosomal protein S30E (Rps30E), Large ribosomal subunit protein eL8, MAGNESIUM ION, ... | | Authors: | Bourgeois, G, Coureux, P.D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2024-06-19 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of Saccharolobus solfataricus initiation complexes with leaderless mRNAs highlight archaeal features and eukaryotic proximity.
Nat Commun, 16, 2025
|
|
9KNX
 
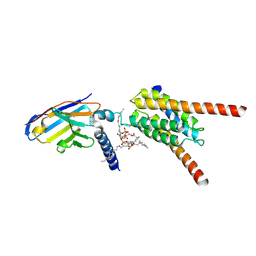 | |
