4YU8
 
 | | Crystal structure of Neuroblastoma suppressor of tumorigenicity 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Neuroblastoma suppressor of tumorigenicity 1 | | Authors: | Ren, J, Nettleship, J.E, Stammers, D.K, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2015-03-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Neuroblastoma suppressor of tumorigenicity 1
To Be Published
|
|
8WQN
 
 | |
7YWP
 
 | | Closed conformation of Oligopeptidase B from Serratia proteomaculans with covalently bound TCK | | Descriptor: | N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, Oligopeptidase B | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Inhibitor-Bound Bacterial Oligopeptidase B in the Closed State: Similarity and Difference between Protozoan and Bacterial Enzymes.
Int J Mol Sci, 24, 2023
|
|
6YM3
 
 | | Crystal structure of Compound 1 with PIP4K2A | | Descriptor: | (2~{R})-2-[[3-cyano-2-[4-(2-ethoxyphenyl)phenyl]-5,8-dihydro-1,7-naphthyridin-4-yl]amino]propanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6Q4S
 
 | | Crystal structure of a-eudesmol synthase | | Descriptor: | CHLORIDE ION, Pentalenene synthase | | Authors: | Correia Cordeiro, R.S, Hakansson, M, Logan, D.T, Kourist, R. | | Deposit date: | 2018-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of three novel sesquiterpene synthases from Streptomyces chartreusis NRRL 3882 and crystal structure of an alpha-eudesmol synthase.
J.Biotechnol., 297, 2019
|
|
6YM4
 
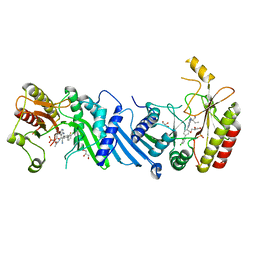 | | Crystal structure of BAY-297 with PIP4K2A | | Descriptor: | (2~{R})-2-[[2-[4-(3-chloranyl-2-fluoranyl-phenyl)phenyl]-3-cyano-1,7-naphthyridin-4-yl]amino]butanamide, GLYCEROL, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
3MQL
 
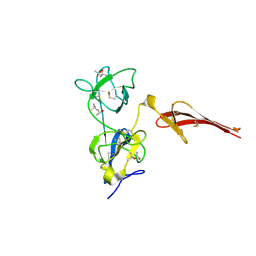 | | Crystal structure of the fibronectin 6FnI1-2FnII7FnI fragment | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Erat, M.C, Campbell, I.D, Vakonakis, I. | | Deposit date: | 2010-04-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Implications for collagen binding from the crystallographic structure of fibronectin 6FnI1-2FnII7FnI
J.Biol.Chem., 285, 2010
|
|
8K56
 
 | |
7KJV
 
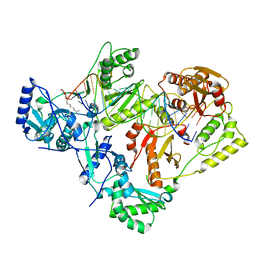 | | Structure of HIV-1 reverse transcriptase initiation complex core | | Descriptor: | HIV-1 viral RNA fragment, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
7STD
 
 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 4 | | Descriptor: | ((1RS,3SR)-2,2-DICHLORO-N-[(R)-1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL-3-METHYLCYCLOPROPANECARBOXAMIDE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-11 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
6YS2
 
 | | Crystal structure of FAP R451A in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRX
 
 | | Low-dose crystal structure of FAP at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
4YS6
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTANS (Cphy_1585, TARGET EFI-511156) WITH BOUND BETA-D-GLUCOSE | | Descriptor: | CHLORIDE ION, Putative solute-binding component of ABC transporter, SODIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTANS (Cphy_1585, TARGET EFI-511156) WITH BOUND BETA-D-GLUCOSE
To be published
|
|
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
8BEX
 
 | | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein at pH 3 | | Descriptor: | Cry49Aa protein | | Authors: | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R. | | Deposit date: | 2022-10-22 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6YRZ
 
 | | Crystal structure of FAP et pH 8.5 after illumination at 150K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, ... | | Authors: | Sorigue, D, Legrand, P, Blangy, S, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
8BEZ
 
 | | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein at pH 11 | | Descriptor: | Cry49Aa protein | | Authors: | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R. | | Deposit date: | 2022-10-22 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1ATI
 
 | | CRYSTAL STRUCTURE OF GLYCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | Descriptor: | GLYCYL-TRNA SYNTHETASE, GLYCYL-tRNA SYNTHETASE | | Authors: | Logan, D.T, Mazauric, M.-H, Kern, D, Moras, D. | | Deposit date: | 1996-04-23 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of glycyl-tRNA synthetase from Thermus thermophilus.
EMBO J., 14, 1995
|
|
6YS1
 
 | | Crystal structure of FAP R451K mutant in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6EXP
 
 | | Crystal structure of the SIRV3 AcrID1 (gp02) anti-CRISPR protein | | Descriptor: | SIRV3 AcrID1 (gp02) anti-CRISPR protein | | Authors: | He, F, Bhoobalan-Chitty, Y, Van, L.B, Kjeldsen, A.L, Dedola, M, Makarova, K.S, Koonin, E.V, Brodersen, D.E, Peng, X. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity.
Nat Microbiol, 3, 2018
|
|
6VVC
 
 | |
5JH7
 
 | | Tubulin-Eribulin complex | | Descriptor: | (1S,3S,6S,9S,12S,14R,16R,18S,20R,21R,22S,26R,29S,31R,32S,33R,35R,36S)-20-[(2S)-3-amino-2-hydroxypropyl]-21-methoxy-14-methyl-8,15-dimethylidene-2,19,30,34,37,39,40,41-octaoxanonacyclo[24.9.2.1~3,32~.1~3,33~.1~6,9~.1~12,16~.0~18,22~.0~29,36~.0~31,35~]hentetracontan-24-one (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Doodhi, H, Prota, A.E, Rodriguez-Garcia, R, Xiao, H, Custar, D.W, Bargsten, K, Katrukha, E.A, Hilbert, M, Hua, S, Jiang, K, Grigoriev, I, Yang, C.-P.H, Cox, D, Band Horwitz, S, Kapitein, L.C, Akhmanova, A, Steinmetz, M.O. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Termination of Protofilament Elongation by Eribulin Induces Lattice Defects that Promote Microtubule Catastrophes.
Curr.Biol., 26, 2016
|
|
7NG7
 
 | | Src kinase bound to eCF506 trapped in inactive conformation | | Descriptor: | 1,2-ETHANEDIOL, Proto-oncogene tyrosine-protein kinase Src, tert-butyl (4-(4-amino-1-(2-(4-(dimethylamino)piperidin-1-yl)ethyl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl)-2-methoxyphenyl)carbamate | | Authors: | Lietha, D, Unciti-Broceta, A. | | Deposit date: | 2021-02-08 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Conformation Selective Mode of Inhibiting SRC Improves Drug Efficacy and Tolerability.
Cancer Res., 81, 2021
|
|
7MYG
 
 | | M. tb Ag85C modified by THL-10d | | Descriptor: | Diacylglycerol acyltransferase, GLYCEROL | | Authors: | Sudasinghe, T.D, Ronning, D.R. | | Deposit date: | 2021-05-21 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Total Synthesis of Tetrahydrolipstatin, Its Derivatives, and Evaluation of Their Ability to Potentiate Multiple Antibiotic Classes against Mycobacterium Species.
Acs Infect Dis., 7, 2021
|
|
