8EJA
 
 | |
5ELF
 
 | | Cholera toxin El Tor B-pentamer in complex with A-pentasaccharide | | Descriptor: | BICINE, CALCIUM ION, Cholera enterotoxin subunit B, ... | | Authors: | Heggelund, J.E, Burschowsky, D, Krengel, U. | | Deposit date: | 2015-11-04 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-Resolution Crystal Structures Elucidate the Molecular Basis of Cholera Blood Group Dependence.
Plos Pathog., 12, 2016
|
|
8EMF
 
 | |
6N7C
 
 | | Structure of the human JAK1 kinase domain with compound 56 | | Descriptor: | GLYCEROL, N-[5-(3-methoxynaphthalen-2-yl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7NSC
 
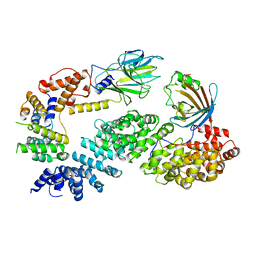 | | Substrate receptor scaffolding module of human CTLH E3 ubiquitin ligase | | Descriptor: | Glucose-induced degradation protein 4 homolog, Glucose-induced degradation protein 8 homolog, Isoform 2 of Armadillo repeat-containing protein 8, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
8EMG
 
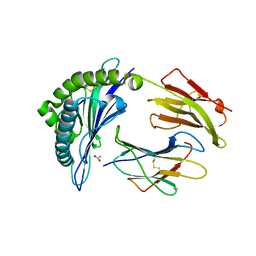 | |
8UF2
 
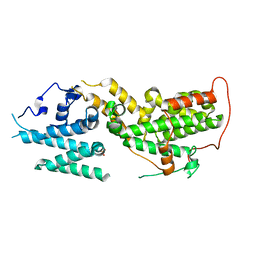 | | Apo SOS2 crystal structure in P1 space group | | Descriptor: | SULFATE ION, Son of sevenless homolog 2 | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
5EN2
 
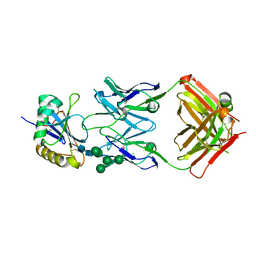 | | Molecular basis for antibody-mediated neutralization of New World hemorrhagic fever mammarenaviruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Mahmutovic, S, Clark, L, Levis, S, Briggiler, A, Enria, D, Harrison, S.C, Abraham, J. | | Deposit date: | 2015-11-09 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Molecular Basis for Antibody-Mediated Neutralization of New World Hemorrhagic Fever Mammarenaviruses.
Cell Host Microbe, 18, 2015
|
|
8EMJ
 
 | |
8EMI
 
 | |
8EMK
 
 | |
8EY1
 
 | |
7NS4
 
 | | Catalytic module of yeast Chelator-GID SR4 E3 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase RMD5, Protein FYV10, ZINC ION | | Authors: | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
6FQJ
 
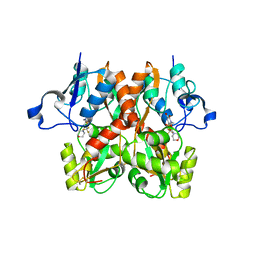 | | GluA2(flop) G724C ligand binding core dimer bound to ZK200775 at 2.50 Angstrom resolution | | Descriptor: | Glutamate receptor 2,Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.500017 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
5NFU
 
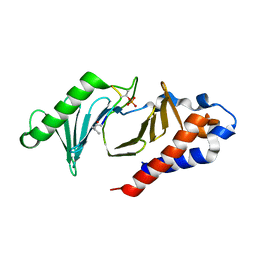 | | The structure of the polo-box domain (PBD) of polo-like kinase 1 (Plk1) in complex with LHSpTA peptide | | Descriptor: | Ac-LEU-HIS-SER-(TPO)-ALA, Serine/threonine-protein kinase PLK1 | | Authors: | Kunciw, D.L, Rossmann, M, De Fusco, C, Spring, D.R, Hyvonen, M. | | Deposit date: | 2017-03-16 | | Release date: | 2018-05-16 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | A cryptic hydrophobic pocket in the polo-box domain of the polo-like kinase PLK1 regulates substrate recognition and mitotic chromosome segregation.
Sci Rep, 9, 2019
|
|
8TVP
 
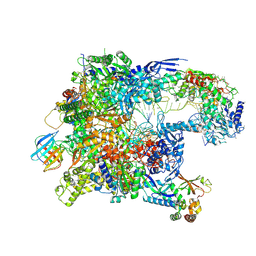 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (open state) | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6SP6
 
 | | Ultra-high Resolution Crystal Structure of the CTX-M-15 Extended-Spectrum beta-Lactamase in Complex with Taniborbactam (VNRX-5133) | | Descriptor: | (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery of Taniborbactam (VNRX-5133): A Broad-Spectrum Serine- and Metallo-beta-lactamase Inhibitor for Carbapenem-Resistant Bacterial Infections.
J.Med.Chem., 63, 2020
|
|
6YCL
 
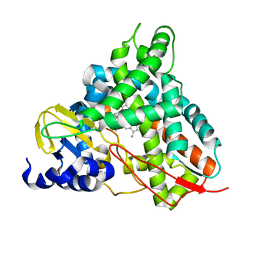 | | Crystal structure of GcoA T296G bound to p-vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
8TVV
 
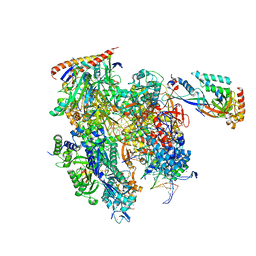 | | Cryo-EM structure of backtracked Pol II | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6YCK
 
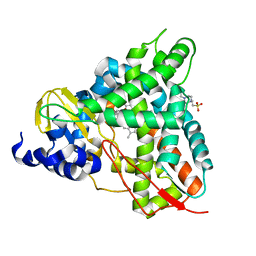 | | Crystal structure of GcoA T296A bound to p-vanillin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
7OB1
 
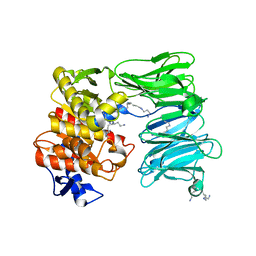 | | OLIGOPEPTIDASE B FROM S. PROTEOMACULANS WITH MODIFIED HINGE | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Korzhenevskiy, D.A, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
6SU7
 
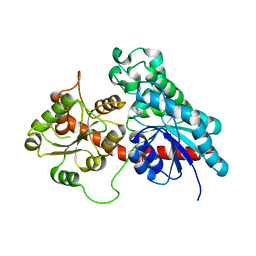 | | Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and 3,4-Dichloroaniline | | Descriptor: | 3,4-Dichloroaniline, Glycosyltransferase | | Authors: | Fredslund, F, Teze, D, Svensson, B, Adams, P.D, Welner, D.H. | | Deposit date: | 2019-09-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | O-/N-/S-Specificity in Glycosyltransferase Catalysis: From Mechanistic Understanding to Engineering
Acs Catalysis, 11, 2021
|
|
6YCO
 
 | | Crystal structure of GcoA F169S bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
5NH6
 
 | |
5JA4
 
 | |
