6ZZB
 
 | |
6OEG
 
 | | Structure of CagX from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagX | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
5IFU
 
 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with Glyburide | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, CHLORIDE ION, ... | | Authors: | Dranow, D.M, Hewitt, S.N, Abendroth, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-26 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Biochemical and Structural Characterization of Selective Allosteric Inhibitors of the Plasmodium falciparum Drug Target, Prolyl-tRNA-synthetase.
ACS Infect Dis, 3, 2017
|
|
8E34
 
 | | CryoEM structures of bAE1 captured in multiple states | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-08-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
7GLQ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with PET-UNK-8df914d1-2 (Mpro-P2007) | | Descriptor: | 2-(3-chlorophenyl)-N-[(4R)-imidazo[1,2-a]pyridin-3-yl]acetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7M7C
 
 | | Crystal Structure of Hip1 (Rv2224c) mutant - T466A/S228DHA (dehydroalanine) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitors and Inactivators of Mycobacterium tuberculosis serine protease Hip1 (Rv2224c)
To Be Published
|
|
6WZM
 
 | | LY3041658 Fab bound to CXCL8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-8, LY3041658 Fab heavy chain, ... | | Authors: | Durbin, J.D, Druzina, Z. | | Deposit date: | 2020-05-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery and characterization of a neutralizing pan-ELR+CXC chemokine monoclonal antibody.
Mabs, 12
|
|
7GM5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-5cd9ea36-2 (Mpro-P2089) | | Descriptor: | (4S)-6-chloro-2-(cyclopropylsulfamoyl)-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6UBE
 
 | | Azide-triggered subtilisin SUBT_BACAM complexed with the peptide LFRAL | | Descriptor: | AZIDE ION, GLYCEROL, Peptide LFRAL, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J, Gallagher, D.T, Custer, G. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
7GMK
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with PET-UNK-d899bab6-1 (Mpro-P2201) | | Descriptor: | 2-(3-chlorophenyl)-N-[6-(dimethylamino)isoquinolin-4-yl]acetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GMW
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-e119ab4f-1 (Mpro-P2224) | | Descriptor: | (4S)-6-chloro-N-(7-fluoroisoquinolin-4-yl)-2-[2-(methylamino)-2-oxoethyl]-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8B9U
 
 | |
7RA7
 
 | | Crystal structure of rabbit anti-HIV Fab 11A | | Descriptor: | 11A Fab heavy chain, 11A Fab light chain, PENTAETHYLENE GLYCOL, ... | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Limited breadth of anti-HIV Env glycan hole antibodies is further hindered by strain-specific peptide interactions
To Be Published
|
|
7GNJ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-976a33d5-1 (Mpro-P2724) | | Descriptor: | 1-[(3'S)-6-chloro-1'-(isoquinolin-4-yl)-1,2'-dioxo-1H-spiro[isoquinoline-4,3'-pyrrolidin]-2(3H)-yl]-N-methylcyclopropane-1-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6LLB
 
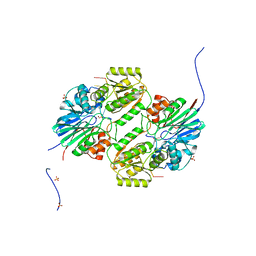 | | Crystal structure of mpy-RNase J (mutant S247A), an archaeal RNase J from Methanolobus psychrophilus R15, in complex with 6 nt RNA | | Descriptor: | MPY-RNase J, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, ... | | Authors: | Li, D.F, Hou, Y.J, Guo, L. | | Deposit date: | 2019-12-22 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A newly identified duplex RNA unwinding activity of archaeal RNase J depends on processive exoribonucleolysis coupled steric occlusion by its structural archaeal loops.
Rna Biol., 17, 2020
|
|
6LYD
 
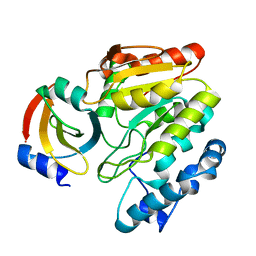 | | Crystal Structure of mimivirus UNG Y322L in complex with UGI | | Descriptor: | Probable uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Pathak, D, Kwon, E, Kim, D.Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Selective interactions between mimivirus uracil-DNA glycosylase and inhibitory proteins determined by a single amino acid.
J.Struct.Biol., 211, 2020
|
|
7P0X
 
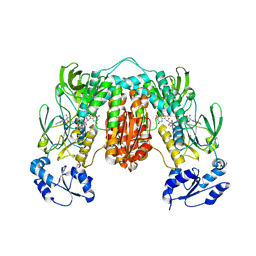 | | Crystal structure of Thioredoxin reductase from Brugia Malayi | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fata, F, Ardini, M, Silvestri, I, Gabriele, F, Cheng, Q, Arner, E.S.J, Williams, D.L. | | Deposit date: | 2021-06-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Biochemical and structural characterizations of thioredoxin reductase selenoproteins of the parasitic filarial nematodes Brugia malayi and Onchocerca volvulus.
Redox Biol, 51, 2022
|
|
6UF4
 
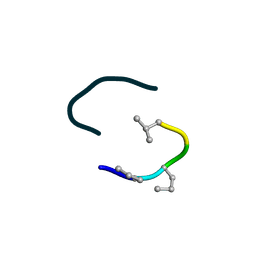 | | S2 symmetric peptide design number 4 crystal form 2, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 2 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6QYL
 
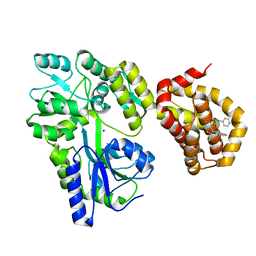 | | Structure of MBP-Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
8BJA
 
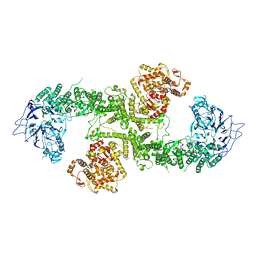 | |
8E8O
 
 | | Cryo-EM structure of human ME3 in the presence of citrate | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malic enzyme, mitochondrial | | Authors: | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
6UG6
 
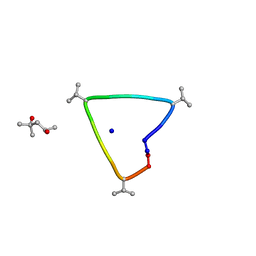 | | C3 symmetric peptide design number 1, Sporty, crystal form 2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, C3-1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6QLV
 
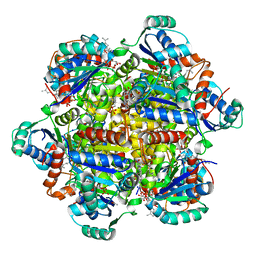 | |
8R6C
 
 | |
6QNB
 
 | |
