7R2V
 
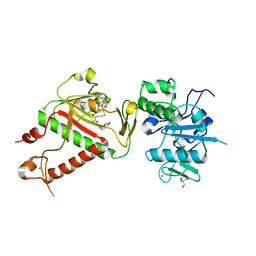 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | Authors: | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | Deposit date: | 2022-02-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
4JZY
 
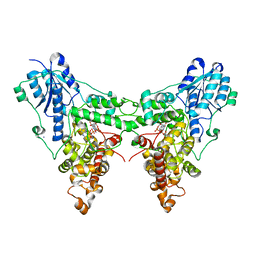 | | Crystal structures of Drosophila Cryptochrome | | Descriptor: | AMMONIUM ION, Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Czarna, A, Wolf, E. | | Deposit date: | 2013-04-03 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function.
Cell(Cambridge,Mass.), 153, 2013
|
|
4K0R
 
 | | Crystal structure of mouse Cryptochrome 1 | | Descriptor: | Cryptochrome-1 | | Authors: | Czarna, A, Wolf, E. | | Deposit date: | 2013-04-04 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function.
Cell(Cambridge,Mass.), 153, 2013
|
|
8QJI
 
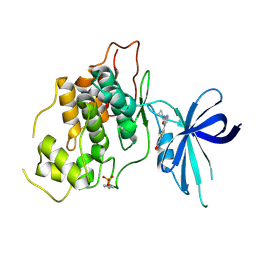 | | Crystal structure of GSK3b in complex with N-(4-(5-(1,2,4-oxadiazol-3-yl)thiophen-2-yl)pyridin-2-yl)cyclopropanecarboxamide inhibitor (TW362) | | Descriptor: | Glycogen synthase kinase-3 beta, N-[4-[5-(1,2,4-oxadiazol-3-yl)thiophen-2-yl]pyridin-2-yl]cyclopropanecarboxamide | | Authors: | Slugocka, E.A, Grygier, P, Wichur, T, Czarna, A, Wieckowska, A. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Exploring Novel GSK-3 beta Inhibitors for Anti-Neuroinflammatory and Neuroprotective Effects: Synthesis, Crystallography, Computational Analysis, and Biological Evaluation.
Acs Chem Neurosci, 15, 2024
|
|
7NT4
 
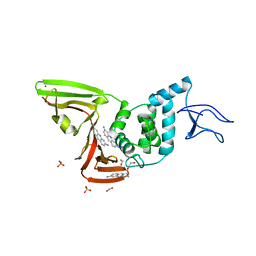 | | X-ray structure of SCoV2-PLpro in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, PROFLAVIN, ... | | Authors: | Napolitano, V, Mourao, A, Bostock, M, Matsuda, A, Czarna, A, Popowicz, G.M. | | Deposit date: | 2021-03-09 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Acriflavine, a clinically approved drug, inhibits SARS-CoV-2 and other betacoronaviruses.
Cell Chem Biol, 29, 2022
|
|
2VID
 
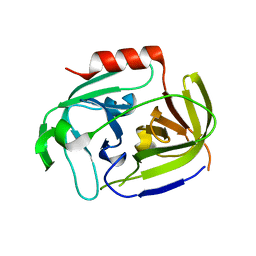 | | Serine protease SplB from Staphylococcus aureus at 1.8A resolution | | Descriptor: | SERINE PROTEASE SPLB | | Authors: | Dubin, G, Stec-Niemczyk, J, Kisielewska, M, Pustelny, K, Popowicz, G.M, Bista, M, Kantyka, T, Boulware, K.T, Stennicke, H.R, Czarna, A, Phopaisarn, M, Daugherty, P.S, Thogersen, I.B, Enghild, J.J, Thornberry, N, Dubin, A, Potempa, J. | | Deposit date: | 2007-11-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzymatic Activity of the Staphylococcus Aureus Splb Serine Protease is Induced by Substrates Containing the Sequence Trp-Glu-Leu-Gln.
J.Mol.Biol., 379, 2008
|
|
7Z1G
 
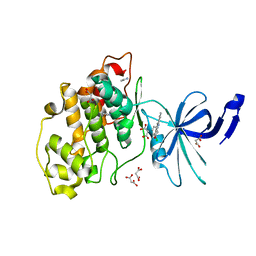 | | Crystal structure of nonphosphorylated (Tyr216) GSK3b in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Glycogen synthase kinase-3 beta, IMIDAZOLE, ... | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-02-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of nonphosphorylated (Tyr216) GSK3b kinase in complex with CX-4945
To Be Published
|
|
7Z1F
 
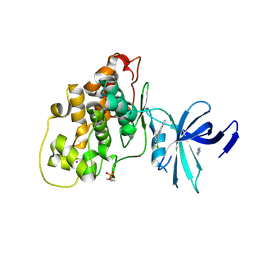 | | Crystal structure of GSK3b in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Glycogen synthase kinase-3 beta, IMIDAZOLE, ... | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-02-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of phosphorylated (Tyr216) GSK3b kinase in complex with CX-4945
To Be Published
|
|
7Z5N
 
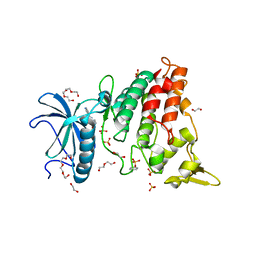 | | Crystal structure of DYRK1A in complex with CX-4945 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pustelny, K, Grygier, P, Golik, P, Dubin, G, Czarna, A. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure DYRK1A in complex with CX-4945
To Be Published
|
|
8C3G
 
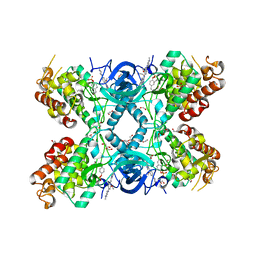 | | Crystal structure of DYRK1A in complex with AZ191 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, N-[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]-4-(1-methylpyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine, ... | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural perspective on the design of selective DYRK1B inhibitors
To Be Published
|
|
8C3Q
 
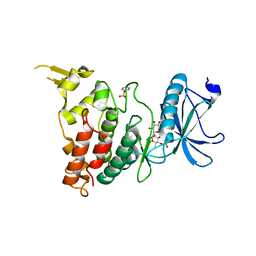 | |
8C3R
 
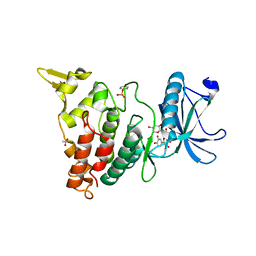 | |
8C2Z
 
 | | Crystal structure of DYRK1B in complex with AZ191 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1B, MANGANESE (II) ION, N-[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]-4-(1-methylpyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural perspective on the design of selective DYRK1B inhibitors
To Be Published
|
|
2Z5S
 
 | | Molecular basis for the inhibition of p53 by Mdmx | | Descriptor: | Cellular tumor antigen p53, Mdm4 protein | | Authors: | Popowicz, G.M, Czarna, A, Rothweiler, U, Szwagierczak, A, Holak, T.A. | | Deposit date: | 2007-07-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of p53 by Mdmx.
Cell Cycle, 6, 2007
|
|
2Z5T
 
 | | Molecular basis for the inhibition of p53 by Mdmx | | Descriptor: | Cellular tumor antigen p53, Mdm4 protein | | Authors: | Popowicz, G.M, Czarna, A, Rothweiler, U, Szwagierczak, A, Holak, T.A. | | Deposit date: | 2007-07-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of p53 by Mdmx.
Cell Cycle, 6, 2007
|
|
3LBL
 
 | | Structure of human MDM2 protein in complex with Mi-63-analog | | Descriptor: | (2'R,3R,4'R,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(2-morpholin-4-ylethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LBK
 
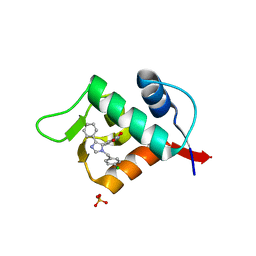 | | Structure of human MDM2 protein in complex with a small molecule inhibitor | | Descriptor: | 6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LBJ
 
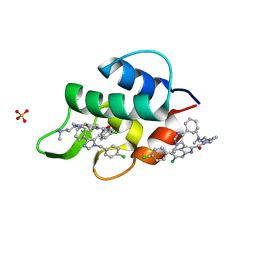 | | Structure of human MDMX protein in complex with a small molecule inhibitor | | Descriptor: | N-[(3S)-1-({6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indol-2-yl}carbonyl)pyrrolidin-3-yl]-N,N',N'-trimethylpropane-1,3-diamine, Protein Mdm4, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3DAC
 
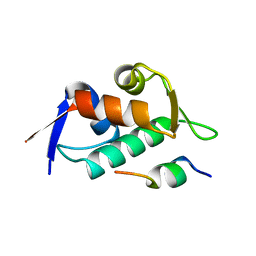 | |
3DAB
 
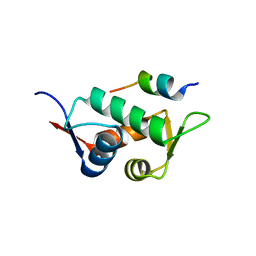 | |
4K1S
 
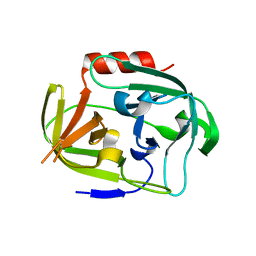 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.96 A resolution | | Descriptor: | Serine protease SplB | | Authors: | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
4K1T
 
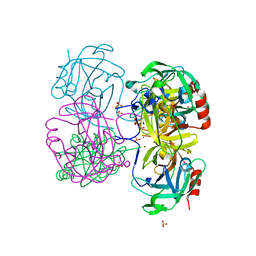 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.60 A resolution | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine protease SplB, ... | | Authors: | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
9EWO
 
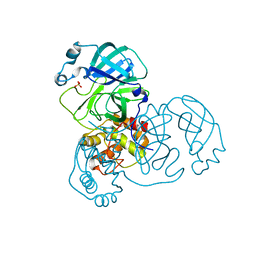 | | Mpro from SARS-CoV-2 with R4A R298A double mutations | | Descriptor: | Non-structural protein 11, SULFATE ION | | Authors: | Plewka, J, Lis, K, Chykunova, Y, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EWM
 
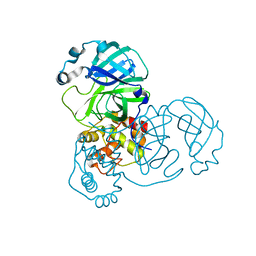 | | Mpro from SARS-CoV-2 with R4Q R298Q double mutations | | Descriptor: | Non-structural protein 11 | | Authors: | Plewka, J, Lis, K, Chykunova, Y, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EWN
 
 | | Mpro from SARS-CoV-2 with 4Q mutation | | Descriptor: | Non-structural protein 11 | | Authors: | Plewka, J, Lis, K, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
