1IJI
 
 | | Crystal Structure of L-Histidinol Phosphate Aminotransferase with PLP | | Descriptor: | Histidinol Phosphate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Li, Y, Larocque, R, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-29 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
1NQC
 
 | | Crystal structures of Cathepsin S inhibitor complexes | | Descriptor: | Cathepsin S, N-[(1R)-2-(BENZYLSULFANYL)-1-FORMYLETHYL]-N-(MORPHOLIN-4-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Pauly, T.A, Sulea, T, Ammirati, M, Sivaraman, J, Danley, D.E, Griffor, M.C, Kamath, A.V, Wang, I.K, Laird, E.R, Menard, R, Cygler, M, Rath, V.L. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specificity determinants of human cathepsin s revealed
by crystal structures of complexes.
Biochemistry, 42, 2003
|
|
1K75
 
 | | The L-histidinol dehydrogenase (hisD) structure implicates domain swapping and gene duplication. | | Descriptor: | GLYCEROL, L-histidinol dehydrogenase, SULFATE ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-10-18 | | Release date: | 2002-02-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KAH
 
 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH L-HISTIDINE (PRODUCT), ZN AND NAD (COFACTOR) | | Descriptor: | HISTIDINE, Histidinol dehydrogenase, ZINC ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KSL
 
 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-01-13 | | Release date: | 2002-04-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1KSV
 
 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URIDINE-5'-MONOPHOSPHATE | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1HBT
 
 | |
1DBO
 
 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-03 | | Release date: | 2000-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
1DBG
 
 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
1GZ0
 
 | | 23S RIBOSOMAL RNA G2251 2'O-METHYLTRANSFERASE RLMB | | Descriptor: | HYPOTHETICAL TRNA/RRNA METHYLTRANSFERASE YJFH | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2002-05-03 | | Release date: | 2002-10-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of the Rlmb 23S Rrna Methyltransferase Reveals a New Methyltransferase Fold with a Unique Knot
Structure, 10, 2002
|
|
1P9K
 
 | | THE SOLUTION STRUCTURE OF YBCJ FROM E. COLI REVEALS A RECENTLY DISCOVERED ALFAL MOTIF INVOLVED IN RNA-BINDING | | Descriptor: | orf, hypothetical protein | | Authors: | Volpon, L, Lievre, C, Osborne, M.J, Gandhi, S, Iannuzzi, P, Larocque, R, Matte, A, Cygler, M, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of YbcJ from Escherichia coli reveals a recently discovered alphaL motif involved in RNA binding.
J.Bacteriol., 185, 2003
|
|
1N3B
 
 | | Crystal Structure of Dephosphocoenzyme A kinase from Escherichia coli | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | O'Toole, N, Barbosa, J.A.R.G, Li, Y, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Trimeric Form of Dephosphocoenzyme A Kinase from Escherichia coli
Protein Sci., 12, 2003
|
|
1FG7
 
 | |
1FG3
 
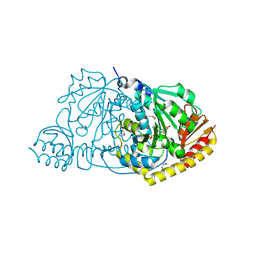 | | CRYSTAL STRUCTURE OF L-HISTIDINOL PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH L-HISTIDINOL | | Descriptor: | HISTIDINOL PHOSPHATE AMINOTRANSFERASE, PHOSPHORIC ACID MONO-[2-AMINO-3-(3H-IMIDAZOL-4-YL)-PROPYL]ESTER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Cygler, M. | | Deposit date: | 2000-07-27 | | Release date: | 2001-08-22 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
1FC5
 
 | |
1F2E
 
 | | STRUCTURE OF SPHINGOMONAD, GLUTATHIONE S-TRANSFERASE COMPLEXED WITH GLUTATHIONE | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Nishio, T, Watanabe, T, Patel, A, Wang, Y, Lau, P.C.K, Grochulski, P, Li, Y, Cygler, M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-06-21 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Properties of a Sphingomonad and Marine Bacterium Beta-Class Glutathione S-Transferases and Crystal Structure of the Former Complex with Glutathione
To be published
|
|
1Q7L
 
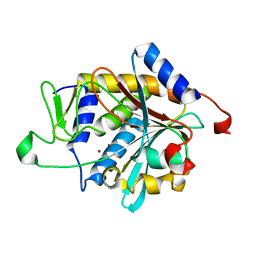 | | Zn-binding domain of the T347G mutant of human aminoacylase-I | | Descriptor: | Aminoacylase-1, GLYCINE, ZINC ION | | Authors: | Lindner, H.A, Lunin, V.V, Alary, A, Hecker, R, Cygler, M, Menard, R. | | Deposit date: | 2003-08-19 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Essential roles of zinc ligation and enzyme dimerization for catalysis in the aminoacylase-1/M20 family.
J.Biol.Chem., 278, 2003
|
|
1DEU
 
 | | CRYSTAL STRUCTURE OF HUMAN PROCATHEPSIN X: A CYSTEINE PROTEASE WITH THE PROREGION COVALENTLY LINKED TO THE ACTIVE SITE CYSTEINE | | Descriptor: | PROCATHEPSIN X | | Authors: | Sivaraman, J, Nagler, D.K, Zhang, R, Menard, R, Cygler, M. | | Deposit date: | 1999-11-15 | | Release date: | 2000-02-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human procathepsin X: a cysteine protease with the proregion covalently linked to the active site cysteine.
J.Mol.Biol., 295, 2000
|
|
1O9B
 
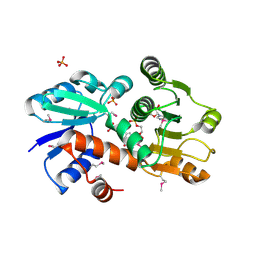 | | QUINATE/SHIKIMATE DEHYDROGENASE YDIB COMPLEXED WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, PHOSPHATE ION | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2002-12-12 | | Release date: | 2003-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Shikimate Dehydrogenase Aroe and its Paralog Ydib: A Common Structural Framework for Different Activities
J.Biol.Chem., 278, 2003
|
|
1PS6
 
 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-HYDROXY-L-THREONINE-5-MONOPHOSPHATE, 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1PS7
 
 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1PTM
 
 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, PHOSPHATE ION, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-06-23 | | Release date: | 2003-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1P9N
 
 | | Crystal structure of Escherichia coli MobB. | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | Rangarajan, S.E, Tocilj, A, Li, Y, Iannuzzi, P, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecules of Escherichia coli MobB assemble into densely packed hollow cylinders in a crystal lattice with 75% solvent content.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1MC3
 
 | | CRYSTAL STRUCTURE OF RFFH | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sivaraman, J, Sauve, V, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-08-05 | | Release date: | 2002-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Escherichia coli Glucose-1-Phosphate Thymidylyltransferase (RffH) Complexed with dTTP and Mg2+
J.BIOL.CHEM., 277, 2002
|
|
1MHW
 
 | | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides | | Descriptor: | 4-biphenylacetyl-Cys-(D)Arg-Tyr-N-(2-phenylethyl) amide, Cathepsin L | | Authors: | Chowdhury, S, Sivaraman, J, Wang, J, Devanathan, G, Lachance, P, Qi, H, Menard, R, Lefebvre, J, Konishi, Y, Cygler, M, Sulea, T, Purisima, E.O. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides
J.Med.Chem., 45, 2002
|
|
