3E6Q
 
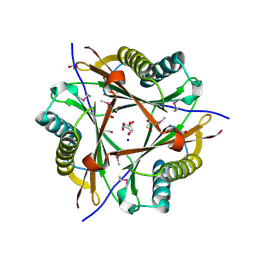 | | Putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-15 | | Release date: | 2008-08-26 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystal structure of putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa.
To be Published
|
|
3E9Q
 
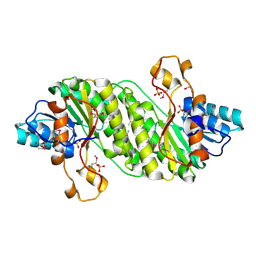 | | Crystal Structure of the Short Chain Dehydrogenase from Shigella flexneri | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-23 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Short Chain Dehydrogenase from Shigella flexneri
To be Published
|
|
3EFG
 
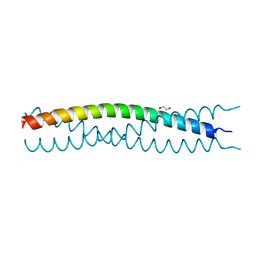 | | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, Protein slyX homolog | | Authors: | Cuff, M.E, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913
TO BE PUBLISHED
|
|
3FG8
 
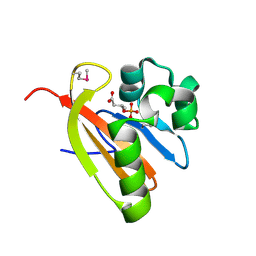 | | Crystal structure of PAS domain of RHA05790 | | Descriptor: | (3R)-3-(phosphonooxy)butanoic acid, uncharacterized protein RHA05790 | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-05 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PAS domain of RHA05790
To be Published
|
|
3F3K
 
 | | The structure of uncharacterized protein YKR043C from Saccharomyces cerevisiae. | | Descriptor: | GLYCEROL, Uncharacterized protein YKR043C | | Authors: | Cuff, M, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
3EY8
 
 | | Structure from the mobile metagenome of V. Pseudocholerae. VPC_CASS1 | | Descriptor: | Biphenyl-2,3-diol 1,2-dioxygenase III-related protein, SULFATE ION | | Authors: | Harrop, S.J, Deshpande, C.N, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-20 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure from the mobile metagenome of V. Pseudocholerae. VPC_CASS1
To be Published
|
|
3F4A
 
 | | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family | | Descriptor: | AMMONIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Osipiuk, J, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family
To be Published
|
|
3CNV
 
 | | Crystal structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica | | Descriptor: | CHLORIDE ION, CITRATE ANION, Putative GntR-family transcriptional regulator | | Authors: | Zimmerman, M.D, Xu, X, Cui, H, Filippova, E.V, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica.
To be Published
|
|
3F5R
 
 | | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p). | | Descriptor: | CHLORIDE ION, FACT complex subunit POB3, FORMIC ACID, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p).
To be Published
|
|
3V75
 
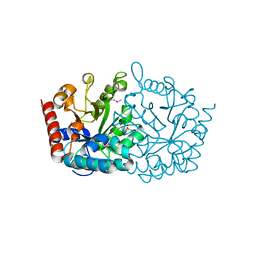 | | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680 | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Stogios, P.J, Xu, X, Cui, H, Kudritska, M, Tan, K, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680
To be Published
|
|
3TTG
 
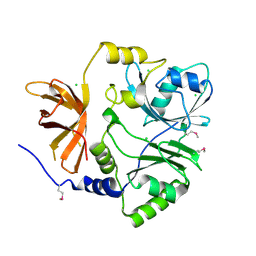 | | Crystal structure of putative aminomethyltransferase from Leptospirillum rubarum | | Descriptor: | CHLORIDE ION, Putative aminomethyltransferase | | Authors: | Michalska, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-14 | | Release date: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative aminomethyltransferase from Leptospirillum rubarum
TO BE PUBLISHED
|
|
3DED
 
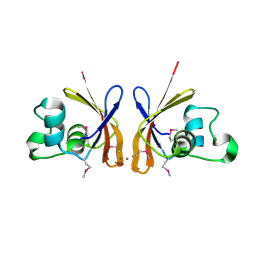 | | C-terminal domain of Probable hemolysin from Chromobacterium violaceum | | Descriptor: | CALCIUM ION, Probable hemolysin | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-09 | | Release date: | 2008-08-05 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of C-terminal domain of Probable hemolysin from Chromobacterium violaceum
To be Published
|
|
3EC6
 
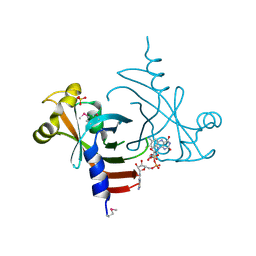 | | Crystal structure of the General Stress Protein 26 from Bacillus anthracis str. Sterne | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, General stress protein 26, SULFATE ION | | Authors: | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the general stress protein 26 from Bacillus anthracis str. Sterne
To be Published
|
|
3EY7
 
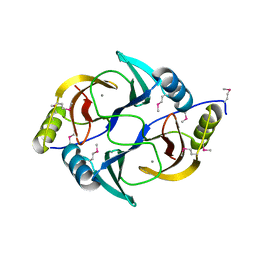 | | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS1 | | Descriptor: | Biphenyl-2,3-diol 1,2-dioxygenase III-related protein, CALCIUM ION | | Authors: | Harrop, S.J, Deshpande, C.N, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-20 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS1
To be Published
|
|
3FHM
 
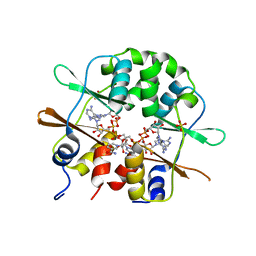 | | Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Zhang, R, Cui, H, Kudritsdka, M, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens
To be Published
|
|
3FBU
 
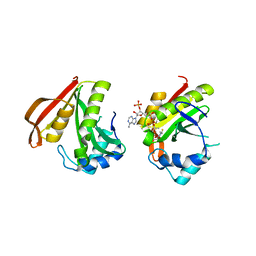 | | The crystal structure of the acetyltransferase (GNAT family) from Bacillus anthracis | | Descriptor: | Acetyltransferase, GNAT family, COENZYME A | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the acetyltransferase (GNAT family) from Bacillus anthracis
To be Published
|
|
3G1J
 
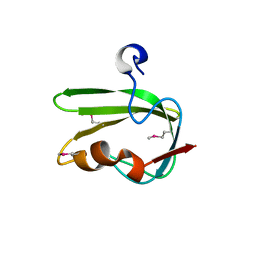 | | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS4. | | Descriptor: | Integron cassette protein | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Tan, K, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS4.
To be published
|
|
3G64
 
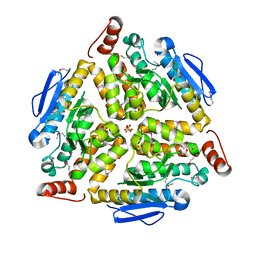 | | Crystal structure of putative enoyl-CoA hydratase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Putative enoyl-CoA hydratase, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Putative Enoyl-CoA Hydratase from Streptomyces coelicolor A3(2)
To be Published
|
|
3GEM
 
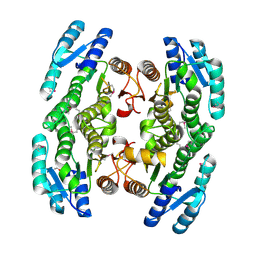 | | Crystal structure of short-chain dehydrogenase from Pseudomonas syringae | | Descriptor: | ACETATE ION, Short chain dehydrogenase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Nocek, B, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray crystal structure of short-chain dehydrogenase from Pseudomonas syringae.
To be Published
|
|
4NNW
 
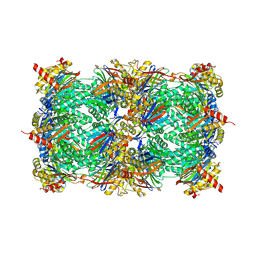 | | yCP in complex with Z-Leu-Leu-Leu-ketoaldehyde | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2R,3S)-1,2-dihydroxy-5-methylhexan-3-yl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3PT1
 
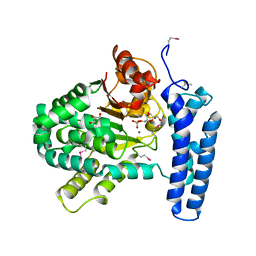 | | Structure of DUF89 from Saccharomyces cerevisiae co-crystallized with F6P. | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Petit, P, Xu, X, Cui, H, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structure and activity of a DUF89 protein from Saccharomyces cerevisiae revealed a novel family of carbohydrate phosphatases
To be Published
|
|
4O8P
 
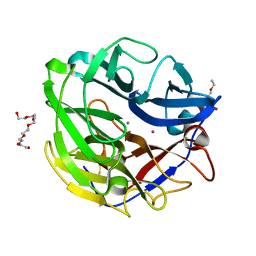 | | Crystal structure of SthAraf62A, a GH62 family alpha-L-arabinofuranosidase from Streptomyces thermoviolaceus, bound to xylotetraose | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Alpha-L-arabinofuranosidase, CALCIUM ION, ... | | Authors: | Stogios, P.J, Wang, W, Xu, X, Cui, H, Master, E, Savchenko, A. | | Deposit date: | 2013-12-28 | | Release date: | 2014-07-02 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.557 Å) | | Cite: | Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 alpha-l-arabinofuranosidase from Streptomyces thermoviolaceus.
Appl.Environ.Microbiol., 80, 2014
|
|
4NO9
 
 | | yCP in complex with Z-Leu-Leu-Leu-epoxyketone | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2R,3S,4S)-1,3-dihydroxy-2,6-dimethylheptan-4-yl]-L-leucinamide, N-[(benzyloxy)carbonyl]-L-leucyl-N-{(1R,2S)-1-hydroxy-4-methyl-1-[(2R)-2-methyloxiran-2-yl]pentan-2-yl}-L-leucinamide, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4MWA
 
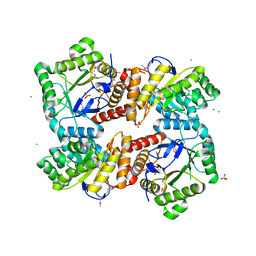 | | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Brunzelle, J.S, Xu, X, Cui, H, Maltseva, N, Bishop, B, Kwon, K, Savchenko, A, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
4NNN
 
 | | yCP in complex with MG132 | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-18 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
