3IO8
 
 | | BimL12F in complex with Bcl-xL | | Descriptor: | Bcl-2-like protein 1, Bcl-2-like protein 11, ZINC ION | | Authors: | Colman, P.M, Lee, E.F, Fairlie, W.D, Smith, B.J, Czabotar, P.E, Yang, H, Sleebs, B.E, Lessene, G. | | Deposit date: | 2009-08-14 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
4CIM
 
 | | Complex of a Bcl-w BH3 mutant with a BH3 domain | | Descriptor: | 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 2 | | Authors: | Colman, P.M, Lee, E.F, Fairlie, W.D. | | Deposit date: | 2013-12-12 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Functional Differences of Pro-Survival and Pro-Apoptotic B Cell Lymphoma 2 (Bcl-2) Proteins Depend on Structural Differences in Their Bcl-2 Homology 3 (Bh3) Domains
J.Biol.Chem., 289, 2014
|
|
4CIN
 
 | | Complex of Bcl-xL with its BH3 domain | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Colman, P.M, Lee, E.F, Fairlie, W.D. | | Deposit date: | 2013-12-12 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | The Functional Differences of Pro-Survival and Pro-Apoptotic B Cell Lymphoma 2 (Bcl-2) Proteins Depend on Structural Differences in Their Bcl-2 Homology 3 (Bh3) Domains
J.Biol.Chem., 289, 2014
|
|
4NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
5NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
2PHL
 
 | | THE STRUCTURE OF PHASEOLIN AT 2.2 ANGSTROMS RESOLUTION: IMPLICATIONS FOR A COMMON VICILIN(SLASH)LEGUMIN STRUCTURE AND THE GENETIC ENGINEERING OF SEED STORAGE PROTEINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHASEOLIN, PHOSPHATE ION | | Authors: | Lawrence, M.C, Izard, T, Beuchat, M, Blagrove, R.J, Colman, P.M. | | Deposit date: | 1994-07-07 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of phaseolin at 2.2 A resolution. Implications for a common vicilin/legumin structure and the genetic engineering of seed storage proteins.
J.Mol.Biol., 238, 1994
|
|
7NN9
 
 | | NATIVE INFLUENZA VIRUS NEURAMINIDASE SUBTYPE N9 (TERN) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Varghese, J.N, Colman, P.M. | | Deposit date: | 1995-03-15 | | Release date: | 1996-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of the complex of 4-guanidino-Neu5Ac2en and influenza virus neuraminidase.
Protein Sci., 4, 1995
|
|
3NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
1V3C
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III: complex with NEU5AC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1V2I
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-16 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1V3B
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
8G1T
 
 | | Crystal structure of Bax core domain BH3-groove dimer - tetrameric fraction P21 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E, Miller, M.S. | | Deposit date: | 2023-02-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
4ZIF
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3mini | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZIG
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BidBH3mini | | Descriptor: | Apoptosis regulator BAX, BH3-interacting domain death agonist | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4ZII
 
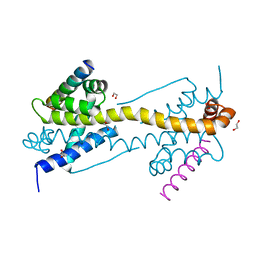 | | Crystal Structure of core/latch dimer of BaxI66A in complex with BidBH3 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, BH3-interacting domain death agonist | | Authors: | Czabotar, P.E, Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
1A14
 
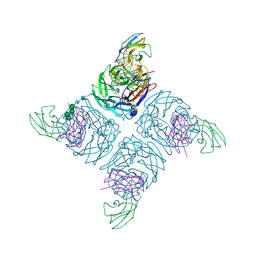 | | COMPLEX BETWEEN NC10 ANTI-INFLUENZA VIRUS NEURAMINIDASE SINGLE CHAIN ANTIBODY WITH A 5 RESIDUE LINKER AND INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NC10 FV (HEAVY CHAIN), ... | | Authors: | Malby, R.L, Mccoy, A.J, Kortt, A.A, Hudson, P.J, Colman, P.M. | | Deposit date: | 1997-12-21 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of single-chain Fv-neuraminidase complexes.
J.Mol.Biol., 279, 1998
|
|
5VWZ
 
 | | Bak in complex with Bim-h3Pc | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, AMMONIUM ION, Bcl-2 homologous antagonist/killer, ... | | Authors: | Brouwer, J.M, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.622 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
5VWY
 
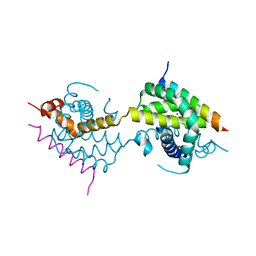 | |
5VX3
 
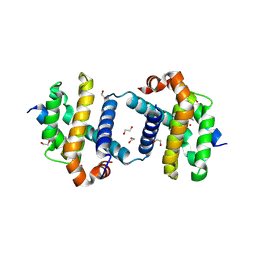 | | Bcl-xL in complex with Bim-h3Pc-RT | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-like protein 1, Bcl-2-like protein 11 | | Authors: | Cowan, A.D, Brouwer, J.M, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
5VWX
 
 | | Bak core latch dimer in complex with Bim-h0-h3Glt | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 11 | | Authors: | Brouwer, J.M, Lan, P, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
4TUH
 
 | | Bcl-xL in complex with inhibitor (Compound 10) | | Descriptor: | 1,2-ETHANEDIOL, 2-[8-(1,3-benzothiazol-2-ylcarbamoyl)-3,4-dihydroisoquinolin-2(1H)-yl]-5-{3-[4-(1H-pyrazolo[3,4-d]pyrimidin-1-yl)phenoxy]propyl}-1,3-thiazole-4-carboxylic acid, ACETATE ION, ... | | Authors: | Czabotar, P.E, Lessense, G, Smith, B.J, Colman, P.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Rescaffolding of Selective Antagonists of BCL-XL.
Acs Med.Chem.Lett., 5, 2014
|
|
5W5X
 
 | | Crystal structure of BAXP168G in complex with an activating antibody | | Descriptor: | 1,2-ETHANEDIOL, 3C10 Fab' heavy chain, 3C10 Fab' light chain, ... | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
4U2V
 
 | | Bak BH3-in-Groove dimer (GFP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Green fluorescent protein,Bcl-2 homologous antagonist/killer | | Authors: | Brouwer, J.M, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bak Core and Latch Domains Separate during Activation, and Freed Core Domains Form Symmetric Homodimers.
Mol.Cell, 55, 2014
|
|
5W62
 
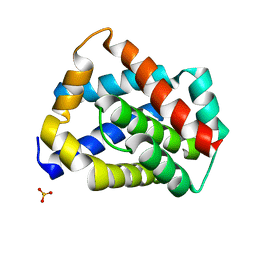 | | Crystal structure of mouse BAX monomer | | Descriptor: | Apoptosis regulator BAX, SULFATE ION | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E, Luo, C.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
5W5Z
 
 | | Crystal structure of BAXP168G in complex with an activating antibody at high resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C10 Fab heavy chain, ... | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
