5O1V
 
 | | Crystal structure of WNK3 kinase domain in a monophosphorylated apo state (A-loop swapped) | | 分子名称: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase WNK3 | | 著者 | Pinkas, D.M, Bufton, J.C, Kupinska, K, Wang, D, Fairhead, M, Kopec, J, Sethi, R, Dixon-Clarke, S.E, Chalk, R, Berridge, G, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | 登録日 | 2017-05-19 | | 公開日 | 2017-06-28 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.723 Å) | | 主引用文献 | Crystal structure of WNK3 kinase domain in a monophosphorylated apo state (A-loop swapped)
To Be Published
|
|
1JG1
 
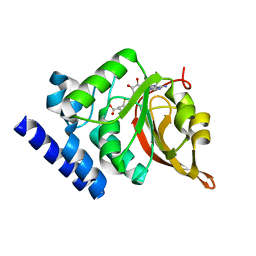 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-ADENOSYL-L-HOMOCYSTEINE | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, protein-L-isoaspartate O-methyltransferase | | 著者 | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | 登録日 | 2001-06-22 | | 公開日 | 2001-11-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG4
 
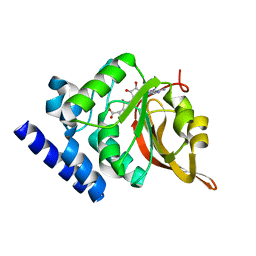 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-adenosylmethionine | | 分子名称: | S-ADENOSYLMETHIONINE, protein-L-isoaspartate O-methyltransferase | | 著者 | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | 登録日 | 2001-06-22 | | 公開日 | 2001-11-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG3
 
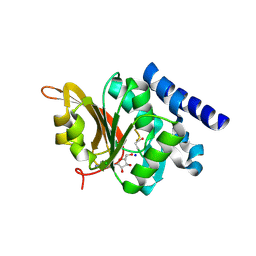 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine & VYP(ISP)HA substrate | | 分子名称: | ADENOSINE, CHLORIDE ION, SODIUM ION, ... | | 著者 | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | 登録日 | 2001-06-22 | | 公開日 | 2001-11-16 | | 最終更新日 | 2011-07-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG2
 
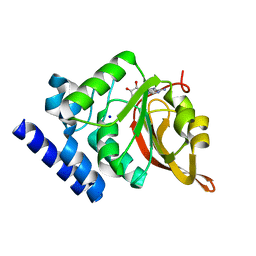 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine | | 分子名称: | ADENOSINE, SODIUM ION, protein-L-isoaspartate O-methyltransferase | | 著者 | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | 登録日 | 2001-06-22 | | 公開日 | 2001-11-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
4UIJ
 
 | | Crystal structure of the BTB domain of KCTD13 | | 分子名称: | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN 1, CHLORIDE ION | | 著者 | Pinkas, D.M, Sanvitale, C.E, Sorell, F.J, Solcan, N, Goubin, S, Canning, P, Williams, E, Chaikuad, A, Dixon Clarke, S.E, Tallant, C, Fonseca, M, Chalk, R, Doutch, J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | 登録日 | 2015-03-30 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
4CKR
 
 | | Crystal structure of the human DDR1 kinase domain in complex with DDR1-IN-1 | | 分子名称: | 1,2-ETHANEDIOL, 4-[(4-ethylpiperazin-1-yl)methyl]-n-{4-methyl-3-[(2-oxo-2,3-dihydro-1h-indol-5-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | 著者 | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Newman, J.A, Dixon-Clarke, S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | 登録日 | 2014-01-07 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of a Potent and Selective Ddr1 Receptor Tyrosine Kinase Inhibitor.
Acs Chem.Biol., 8, 2013
|
|
5FII
 
 | | Structure of a human aspartate kinase, chorismate mutase and TyrA domain. | | 分子名称: | PHENYLALANINE, PHENYLALANINE-4-HYDROXYLASE | | 著者 | Patel, D, Kopec, J, Shrestha, L, Fitzpatrick, F, Pinkas, D, Chaikuad, A, Dixon-Clarke, S, McCorvie, T.J, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | 登録日 | 2015-09-25 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis for Ligand-Dependent Dimerization of Phenylalanine Hydroxylase Regulatory Domain.
Sci.Rep., 6, 2016
|
|
5A15
 
 | | Crystal structure of the BTB domain of human KCTD16 | | 分子名称: | BTB/POZ DOMAIN-CONTAINING PROTEIN KCTD16 | | 著者 | Pinkas, D.M, Sanvitale, C.E, Solcan, N, Goubin, S, Canning, P, Dixon Clarke, S.E, Talon, R, Wiggers, H.J, Fitzpatrick, F, Tallant, C, Kopec, J, Chalk, R, Doutch, J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | 登録日 | 2015-04-28 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
3JQ3
 
 | | Crystal Structure of Lombricine Kinase, complexed with substrate ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lombricine kinase | | 著者 | Bush, D.J, Kirillova, O, Clark, S.A, Fabiola, F, Somasundaram, T, Chapman, M.S. | | 登録日 | 2009-09-05 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.503 Å) | | 主引用文献 | The structure of lombricine kinase: implications for phosphagen kinase conformational changes.
J.Biol.Chem., 286, 2011
|
|
7KFV
 
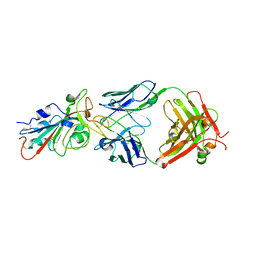 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-B12 Fab) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody C1A-B12 Fab, Spike glycoprotein, ... | | 著者 | Pan, J, Abraham, J, Clark, L, Clark, S. | | 登録日 | 2020-10-15 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFW
 
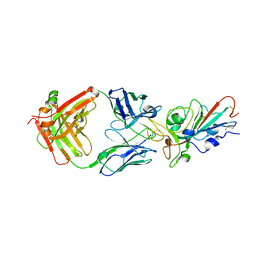 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-B3 Fab) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of antibody C1A-B3 Fab, ... | | 著者 | Pan, J, Abraham, J, Clark, L, Clark, S. | | 登録日 | 2020-10-15 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.792 Å) | | 主引用文献 | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7KFY
 
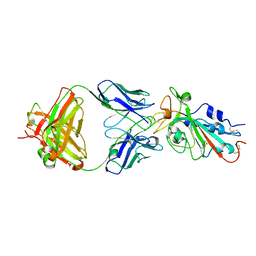 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-F10 Fab) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of human antibody C1A-F10 Fab, ... | | 著者 | Pan, J, Abraham, J, Clark, L, Clark, S. | | 登録日 | 2020-10-15 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.157 Å) | | 主引用文献 | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
7SN1
 
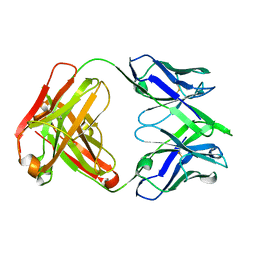 | |
7SN0
 
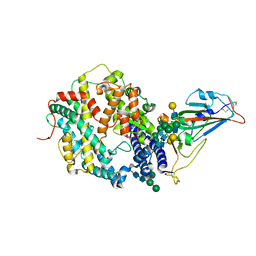 | | Crystal structure of spike protein receptor binding domain of escape mutant SARS-CoV-2 from immunocompromised patient (d146*) in complex with human receptor ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pan, J, Abraham, J, Clark, S. | | 登録日 | 2021-10-27 | | 公開日 | 2021-12-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7KFX
 
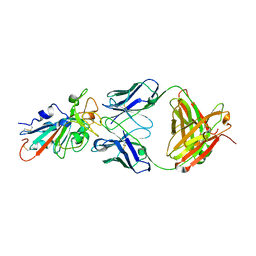 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-C2 Fab) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of human antibody C1A-C2 Fab, ... | | 著者 | Pan, J, Abraham, J, Clark, L, Clark, S. | | 登録日 | 2020-10-15 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.226 Å) | | 主引用文献 | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
4LMQ
 
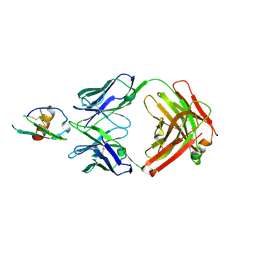 | | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12 | | 分子名称: | Stromal cell-derived factor 1, hu30D8 Fab heavy chain, hu30D8 Fab light chain | | 著者 | Zhong, Z, Wang, J, Li, B, Xiang, H, Ultsch, M, Coons, M, Wong, T, Chiang, N.Y, Clark, S, Clark, R, Quintana, L, Gribling, P, Suto, E, Barck, K, Corpuz, R, Yao, J, Takkar, R, Lee, W.P, Damico-Beyer, L.A, Carano, R.D, Adams, C, Kelley, R.F, Wang, W, Ferrara, N. | | 登録日 | 2013-07-10 | | 公開日 | 2013-08-14 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.773 Å) | | 主引用文献 | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12.
Clin.Cancer Res., 19, 2013
|
|
7USY
 
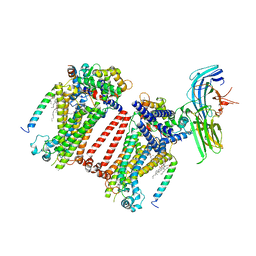 | | Structure of C. elegans TMC-1 complex with ARRD-6 | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Jeong, H, Clark, S, Gouaux, E. | | 登録日 | 2022-04-26 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
7USW
 
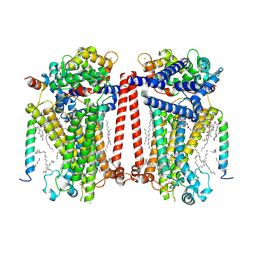 | | Structure of Expanded C. elegans TMC-1 complex | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Jeong, H, Clark, S, Gouaux, E. | | 登録日 | 2022-04-26 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
7USX
 
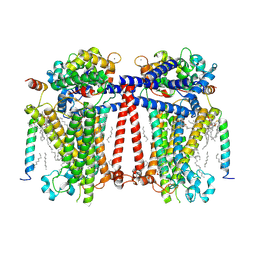 | | Structure of Contracted C. elegans TMC-1 complex | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Jeong, H, Clark, S, Gouaux, E. | | 登録日 | 2022-04-26 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
1P52
 
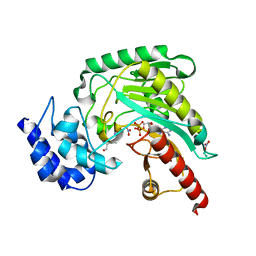 | | Structure of Arginine kinase E314D mutant | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, D-ARGININE, ... | | 著者 | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | 登録日 | 2003-04-24 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
1P50
 
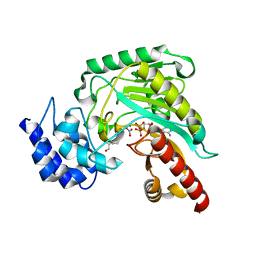 | | Transition state structure of an Arginine Kinase mutant | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | 著者 | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | 登録日 | 2003-04-24 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
2UWN
 
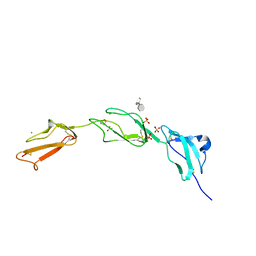 | | Crystal structure of Human Complement Factor H, SCR domains 6-8 (H402 risk variant), in complex with ligand. | | 分子名称: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | 著者 | Prosser, B.E, Johnson, S, Roversi, P, Herbert, A.P, Blaum, B.S, Tyrrell, J, Jowitt, T.A, Clark, S.J, Terelli, E, Uhrin, D, Barlow, P.N, Sim, R.B, Day, A.J, Lea, S.M. | | 登録日 | 2007-03-22 | | 公開日 | 2007-10-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural Basis for Complement Factor H Linked Age-Related Macular Degeneration.
J.Exp.Med., 204, 2007
|
|
2V8E
 
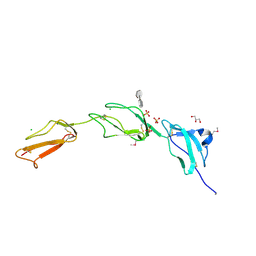 | | Crystal structure of Human Complement Factor H, SCR domains 6-8 (H402 risk variant), in complex with ligand. | | 分子名称: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, CHLORIDE ION, COMPLEMENT FACTOR H, ... | | 著者 | Prosser, B.E, Johnson, S, Roversi, P, Herbert, A.P, Blaum, B.S, Tyrrell, J, Jowitt, T.A, Clark, S.J, Tarelli, E, Uhrin, D, Barlow, P.N, Sim, R.B, Day, A.J, Lea, S.M. | | 登録日 | 2007-08-07 | | 公開日 | 2007-10-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for Complement Factor H Linked Age-Related Macular Degeneration.
J.Exp.Med., 204, 2007
|
|
7ABL
 
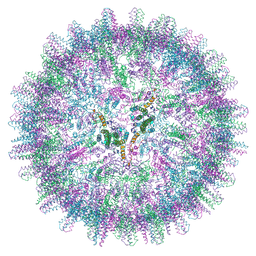 | | HBV pgRNA T=4 NCP icosahedral symmetry | | 分子名称: | Capsid protein | | 著者 | Patel, N, Clark, S, Weis, E.U, Mata, C.P, Bohon, J, Farquhar, E, Ranson, N.A, Twarock, R, Stockley, P.G. | | 登録日 | 2020-09-07 | | 公開日 | 2021-10-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | In vitro functional analysis of gRNA sites regulating assembly of hepatitis B virus.
Commun Biol, 4, 2021
|
|
