2LJV
 
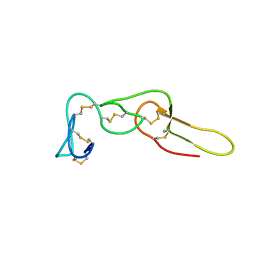 | | Solution structure of Rhodostomin G50L mutant | | Descriptor: | Disintegrin rhodostomin | | Authors: | Chuang, W, Shiu, J, Chen, C, Chen, Y, Chang, Y, Huang, C. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design of Integrin AlphaVbeta3-Specific Disintegrin for Cancer Therapy
To be Published
|
|
1W2I
 
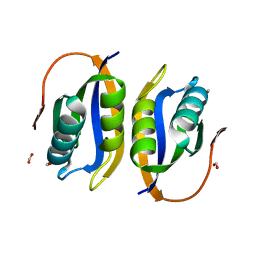 | | Crystal structuore of acylphosphatase from Pyrococcus horikoshii complexed with formate | | Descriptor: | ACYLPHOSPHATASE, FORMIC ACID | | Authors: | Cheung, Y.Y, Lam, S.Y, Chu, W.K, Allen, M.D, Bycroft, M, Wong, K.B. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Hyperthermophilic Archaeal Acylphosphatase from Pyrococcus Horikoshii-Structural Insights Into Enzymatic Catalysis, Thermostability, and Dimerization
Biochemistry, 44, 2005
|
|
209D
 
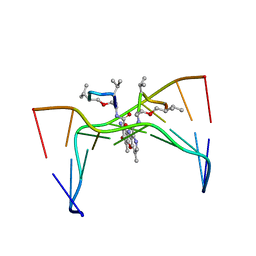 | | Structural, physical and biological characteristics of RNA:DNA binding agent N8-actinomycin D | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3'), N8-ACTINOMYCIN D | | Authors: | Shinomiya, M, Chu, W, Carlson, R.G, Weaver, R.F, Takusagawa, F. | | Deposit date: | 1995-05-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural, Physical, and Biological Characteristics of RNA.DNA Binding Agent N8-Actinomycin D.
Biochemistry, 34, 1995
|
|
2D55
 
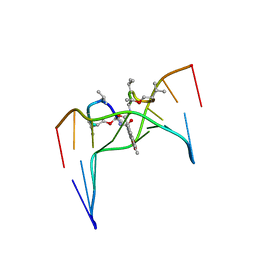 | | Structural, physical and biological characteristics of RNA.DNA binding agent N8-actinomycin D | | Descriptor: | ACTINOMYCIN D, DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3') | | Authors: | Shinomiya, M, Chu, W, Carlson, R.G, Weaver, R.F, Takusagawa, F. | | Deposit date: | 1995-05-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the 2:1 Complex between D(Gaagcttc) and the Anticancer Drug Actinomycin D.
J.Mol.Biol., 225, 1992
|
|
1W5P
 
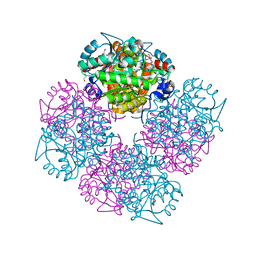 | | Stepwise introduction of zinc binding site into porphobilinogen synthase of Pseudomonas aeruginosa (mutations A129C, D131C, D139C, P132E) | | Descriptor: | 1,2-ETHANEDIOL, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, FORMIC ACID, ... | | Authors: | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | Deposit date: | 2004-08-09 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
1W56
 
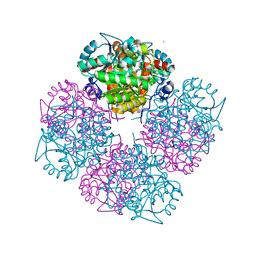 | | Stepwise introduction of zinc binding site into porphobilinogen synthase of Pseudomonas aeruginosa (mutations A129C and D131C) | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | Deposit date: | 2004-08-05 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
1W5M
 
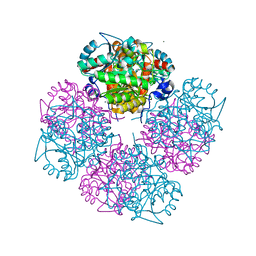 | | Stepwise introduction of zinc binding site into porphobilinogen synthase of Pseudomonas aeruginosa (mutations A129C and D139C) | | Descriptor: | CHLORIDE ION, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, FORMIC ACID, ... | | Authors: | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | Deposit date: | 2004-08-09 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
1W5N
 
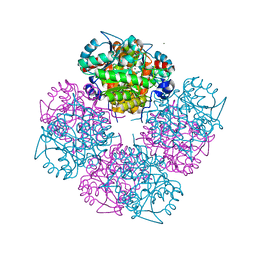 | | Stepwise introduction of zinc binding site into porphobilinogen synthase of Pseudomonas aeruginosa (mutations D131C and D139C) | | Descriptor: | CHLORIDE ION, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, FORMIC ACID, ... | | Authors: | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | Deposit date: | 2004-08-09 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
1W5Q
 
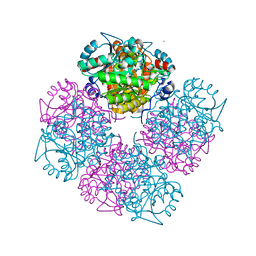 | | Stepwise introduction of zinc binding site into porphobilinogen synthase of Pseudomonas aeruginosa (mutations A129C, D131C, D139C, P132E, K229R) | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | Deposit date: | 2004-08-09 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
1W6T
 
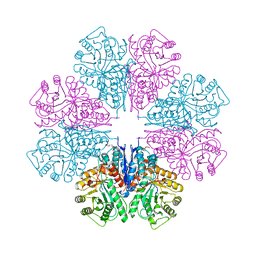 | | Crystal Structure Of Octameric Enolase From Streptococcus pneumoniae | | Descriptor: | ENOLASE, MAGNESIUM ION, NONAETHYLENE GLYCOL | | Authors: | Ehinger, S, Schubert, W.-D, Bergmann, S, Hammerschmidt, S, Heinz, D.W. | | Deposit date: | 2004-08-24 | | Release date: | 2005-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasmin(Ogen)-Binding Alpha-Enolase from Streptococcus Pneumoniae: Crystal Structure and Evaluation of Plasmin(Ogen)-Binding Sites
J.Mol.Biol., 343, 2004
|
|
1W54
 
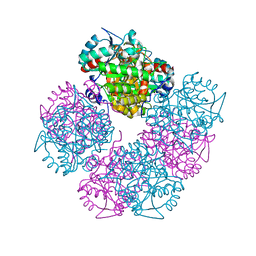 | | Stepwise introduction of a zinc binding site into Porphobilinogen synthase from Pseudomonas aeruginosa (mutation D139C) | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | Deposit date: | 2004-08-05 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
1W5O
 
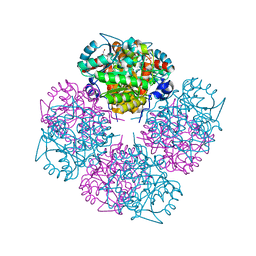 | | Stepwise introduction of zinc binding site into porphobilinogen synthase of Pseudomonas aeruginosa (mutations A129C, D131C and D139C) | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | Deposit date: | 2004-08-09 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
3U8I
 
 | | Functionally selective inhibition of Group IIA phospholipase A2 reveals a role for vimentin in regulating arachidonic acid metabolism | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phospholipase A2, ... | | Authors: | Lee, L.K, Bryant, K.J, Bouveret, R, Lei, P.-W, Duff, A.P, Harrop, S.J, Huang, E.P, Harvey, R.P, Gelb, M.H, Gray, P.P, Curmi, P.M, Cunningham, A.M, Church, W.B, Scott, K.F. | | Deposit date: | 2011-10-17 | | Release date: | 2012-10-17 | | Last modified: | 2013-06-12 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Selective Inhibition of Human Group IIA-secreted Phospholipase A2 (hGIIA) Signaling Reveals Arachidonic Acid Metabolism Is Associated with Colocalization of hGIIA to Vimentin in Rheumatoid Synoviocytes.
J.Biol.Chem., 288, 2013
|
|
3U8H
 
 | | Functionally selective inhibition of Group IIA phospholipase A2 reveals a role for vimentin in regulating arachidonic acid metabolism | | Descriptor: | (S)-5-(4-BENZYLOXY-PHENYL)-4-(7-PHENYL-HEPTANOYLAMINO)-PENTANOIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lee, L.K, Bryant, K.J, Bouveret, R, Lei, P.-W, Duff, A.P, Harrop, S.J, Huang, E.P, Harvey, R.P, Gelb, M.H, Gray, P.P, Curmi, P.M, Cunningham, A.M, Church, W.B, Scott, K.F. | | Deposit date: | 2011-10-17 | | Release date: | 2012-10-17 | | Last modified: | 2013-06-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selective Inhibition of Human Group IIA-secreted Phospholipase A2 (hGIIA) Signaling Reveals Arachidonic Acid Metabolism Is Associated with Colocalization of hGIIA to Vimentin in Rheumatoid Synoviocytes.
J.Biol.Chem., 288, 2013
|
|
3U8D
 
 | | Functionally selective inhibition of Group IIA phospholipase A2 reveals a role for vimentin in regulating arachidonic acid metabolism | | Descriptor: | (3-{[3-(2-amino-2-oxoethyl)-1-benzyl-2-ethyl-1H-indol-5-yl]oxy}propyl)phosphonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lee, L.K, Bryant, K.J, Bouveret, R, Lei, P.-W, Duff, A.P, Harrop, S.J, Huang, E.P, Harvey, R.P, Gelb, M.H, Gray, P.P, Curmi, P.M, Cunningham, A.M, Church, W.B, Scott, K.F. | | Deposit date: | 2011-10-16 | | Release date: | 2012-10-17 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Selective Inhibition of Human Group IIA-secreted Phospholipase A2 (hGIIA) Signaling Reveals Arachidonic Acid Metabolism Is Associated with Colocalization of hGIIA to Vimentin in Rheumatoid Synoviocytes.
J.Biol.Chem., 288, 2013
|
|
1I4A
 
 | | CRYSTAL STRUCTURE OF PHOSPHORYLATION-MIMICKING MUTANT T6D OF ANNEXIN IV | | Descriptor: | ANNEXIN IV, CALCIUM ION, SULFATE ION | | Authors: | Kaetzel, M.A, Mo, Y.D, Mealy, T.R, Campos, B, Bergsma-Schutter, W, Brisson, A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 2001-02-20 | | Release date: | 2001-04-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation mutants elucidate the mechanism of annexin IV-mediated membrane aggregation.
Biochemistry, 40, 2001
|
|
1I7A
 
 | | EVH1 DOMAIN FROM MURINE HOMER 2B/VESL 2 | | Descriptor: | CITRATE ANION, HOMER 2B, PHE-ALA-PHE, ... | | Authors: | Barzik, M, Carl, U.D, Schubert, W.-D, Wehland, J, Heinz, D.W. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The N-terminal domain of Homer/Vesl is a new class II EVH1 domain.
J.Mol.Biol., 309, 2001
|
|
4U5M
 
 | | Structure of a left-handed DNA G-quadruplex | | Descriptor: | DNA (28-MER), MAGNESIUM ION, POTASSIUM ION | | Authors: | Schmitt, E, Mechulam, Y, Phan, A.T, Brahim, H, Chung, W.J, Lim, K.W. | | Deposit date: | 2014-07-25 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a left-handed DNA G-quadruplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3UCI
 
 | | Crystal structure of Rhodostomin ARLDDL mutant | | Descriptor: | disintegrin | | Authors: | Shiu, J.H, Chen, C.Y, Chen, Y.C, Chang, Y.T, Chang, Y.S, Huang, C.H, Chuang, W.J. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design of Integrin AlphaVbeta3-Specific Disintegrin for Cancer Therapy
To be Published
|
|
4WLJ
 
 | | High resolution crystal structure of human kynurenine aminotransferase-I in complex with aminooxyacetate | | Descriptor: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | High resolution crystal structures of human kynurenine aminotransferase-I bound to PLP cofactor, and in complex with aminooxyacetate.
Protein Sci., 26, 2017
|
|
4WP0
 
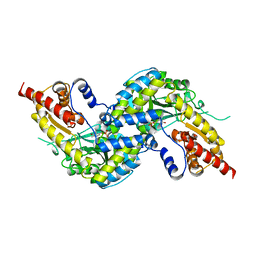 | | Crystal structure of human kynurenine aminotransferase-I with a C-terminal V5-hexahistidine tag | | Descriptor: | Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human kynurenine aminotransferase-I in a novel space group
To be published
|
|
7UWB
 
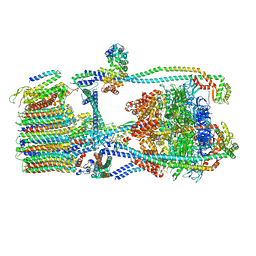 | | Citrus V-ATPase State 2, Highest-Resolution Class | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit AP1 fragment, V-type proton ATPase subunit AP2 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
7UW9
 
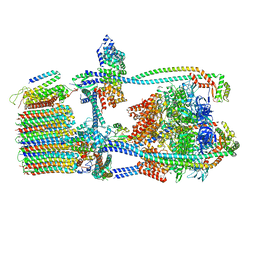 | | Citrus V-ATPase State 1, H in contact with subunit a | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit AP1 fragment, V-type proton ATPase subunit AP2 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
7UWD
 
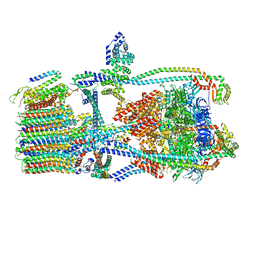 | | Citrus V-ATPase State 2, H in contact with subunits AB | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit, V-type proton ATPase subunit AP1 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
7UWA
 
 | | Citrus V-ATPase State 1, H in contact with subunits AB | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit AP1 fragment, V-type proton ATPase subunit AP2 fragment, ... | | Authors: | Abdelaziz, R.A, Keon, K.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
