2R68
 
 | |
5M1U
 
 | |
5MAA
 
 | | PCE reductive dehalogenase from S. multivorans in complex with 3-bromophenol | | Descriptor: | 3-bromophenol, 5-Methoxybenzimidazolyl-norcobamide, BENZAMIDINE, ... | | Authors: | Kunze, C, Bommer, M, Hagen, W.R, Uksa, M, Dobbek, H, Schubert, T, Diekert, G. | | Deposit date: | 2016-11-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer.
Nat Commun, 8, 2017
|
|
8CPE
 
 | |
5MA0
 
 | | PCE reductive dehalogenase from S. multivorans in complex with 2,6-dichlorophenol | | Descriptor: | 2,6-dichlorophenol, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Kunze, C, Bommer, M, Hagen, W.R, Uksa, M, Dobbek, H, Schubert, T, Diekert, G. | | Deposit date: | 2016-11-02 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer.
Nat Commun, 8, 2017
|
|
5MA1
 
 | | PCE reductive dehalogenase from S. multivorans in complex with 2,4,6-trichlorophenol | | Descriptor: | 2,4,6-trichlorophenol, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Kunze, C, Bommer, M, Hagen, W.R, Uksa, M, Dobbek, H, Schubert, T, Diekert, G. | | Deposit date: | 2016-11-02 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer.
Nat Commun, 8, 2017
|
|
7AWH
 
 | | Crystal structure of human butyrylcholinesterase in complex with tert-butyl 3-(((2-((1-(benzenesulfonyl)-1H-indol-4-yl)oxy)ethyl)amino)methyl)piperidine-1-carboxylate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Wichur, T, Wieckowska, A. | | Deposit date: | 2020-11-08 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development and crystallography-aided SAR studies of multifunctional BuChE inhibitors and 5-HT 6 R antagonists with beta-amyloid anti-aggregation properties.
Eur.J.Med.Chem., 225, 2021
|
|
8DIK
 
 | |
5OBP
 
 | | PCE reductive dehalogenase from S. multivorans with 6-hydroxybenzimidazolyl norcobamide cofactor | | Descriptor: | 6-hydroxybenzimidazolyl-norcobamide, BENZAMIDINE, GLYCEROL, ... | | Authors: | Keller, S, Kunze, C, Bommer, M, Paetz, C, Menezes, R.C, Svatos, A, Dobbek, H, Schubert, T. | | Deposit date: | 2017-06-28 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Selective Utilization of Benzimidazolyl-Norcobamides as Cofactors by the Tetrachloroethene Reductive Dehalogenase of Sulfurospirillum multivorans.
J. Bacteriol., 200, 2018
|
|
4IRZ
 
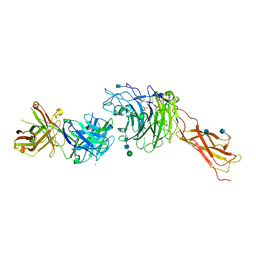 | | Crystal structure of A4b7 headpiece complexed with Fab Natalizumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yu, Y, Schurpf, T, Springer, T.A. | | Deposit date: | 2013-01-15 | | Release date: | 2013-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | How natalizumab binds and antagonizes alpha 4 integrins.
J.Biol.Chem., 288, 2013
|
|
2R60
 
 | |
5JZS
 
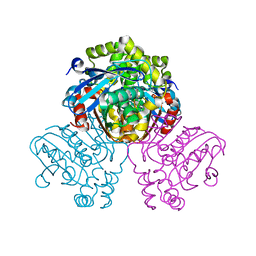 | | HsaD bound to 3,5-dichloro-4-hydroxybenzoic acid | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD, 3,5-dichloro-4-hydroxybenzoic acid | | Authors: | Ryan, A, Polycarpou, E, Lack, N, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E.D, Ballet, R, Abuhammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5JZB
 
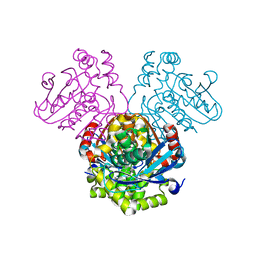 | | Crystal structure of HsaD bound to 3,5-dichlorobenzene sulphonamide | | Descriptor: | 3,5-dichlorobenzene-1-sulfonamide, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, PHOSPHATE ION | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5JZ9
 
 | | Crystal structure of HsaD bound to 3,5-dichloro-4-hydroxybenzenesulphonic acid | | Descriptor: | 3,5-dichloro-4-hydroxybenzene-1-sulfonic acid, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
6CEQ
 
 | |
6MI6
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH AN ADP ANALOG | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy{[(1-hydroxy-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl]disulfanyl}phosphoryl]oxy}phosphoryl]adenosine, ADENOSINE-5'-DIPHOSPHATE, Chemotaxis protein CheA, ... | | Authors: | Crane, B.R, Muok, A.R, Chua, T.K, Le, H. | | Deposit date: | 2018-09-19 | | Release date: | 2018-12-05 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Nucleotide Spin Labeling for ESR Spectroscopy of ATP-Binding Proteins.
Appl.Magn.Reson., 49, 2018
|
|
1PZ7
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1Q56
 
 | | NMR structure of the B0 isoform of the agrin G3 domain in its Ca2+ bound state | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-08-06 | | Release date: | 2004-04-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding
Structure, 12, 2004
|
|
1PZ8
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1PZ9
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
7KH8
 
 | | Human LMPTP in complex with inhibitor | | Descriptor: | 3-[(2,6-dichlorophenyl)methyl]-8-(2-methylphenyl)-3H-purin-6-amine, Low molecular weight phosphotyrosine protein phosphatase, NITRATE ION | | Authors: | Stanford, S.M, Diaz, M.A, Ardecky, R.J, Zou, J, Roosild, T, Holmes, Z.J, Hedrick, M.P, Rodiles, S, Santelli, E, Chung, T.D.Y, Jackson, M.R, Bottini, N, Pinkerton, A.B. | | Deposit date: | 2020-10-20 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Orally Bioavailable Purine-Based Inhibitors of the Low-Molecular-Weight Protein Tyrosine Phosphatase.
J.Med.Chem., 64, 2021
|
|
4APC
 
 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1 | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
4B9D
 
 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) with inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1, ... | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Canning, P, von Delft, F, Raynor, J, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
5BQ1
 
 | | Capturing Carbon Dioxide in beta Carbonic Anhydrase | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase, ZINC ION | | Authors: | Aggarwal, M, Chua, T.K, Pinard, M.A, Szebenyi, D.M, McKenna, R. | | Deposit date: | 2015-05-28 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Carbon Dioxide "Trapped" in a beta-Carbonic Anhydrase.
Biochemistry, 54, 2015
|
|
1FBM
 
 | | ASSEMBLY DOMAIN OF CARTILAGE OLIGOMERIC MATRIX PROTEIN IN COMPLEX WITH ALL-TRANS RETINOL | | Descriptor: | PROTEIN (CARTILAGE OLIGOMERIC MATRIX PROTEIN), RETINOL | | Authors: | Guo, Y, Bozic, D, Malashkevich, V.N, Kammerer, R.A, Schulthess, T. | | Deposit date: | 2000-07-16 | | Release date: | 2000-08-02 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | All-trans retinol, vitamin D and other hydrophobic compounds bind in the axial pore of the five-stranded coiled-coil domain of cartilage oligomeric matrix protein.
EMBO J., 17, 1998
|
|
