1JLR
 
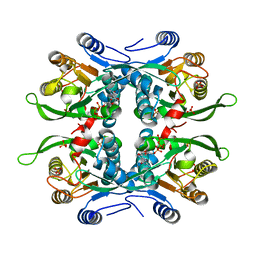 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE GTP COMPLEX 2 MUTANT C128V | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, Uracil Phosphoribosyltransferase | | Authors: | Schumacher, M.A, Bashor, C.J, Otsu, K, Zu, S, Parry, R, Ulmman, B, Brennan, R.G. | | Deposit date: | 2001-07-16 | | Release date: | 2002-01-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7LQ2
 
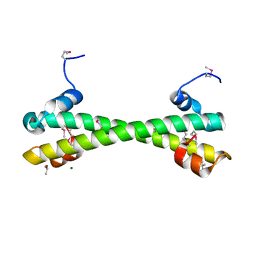 | | Apo Rr RsiG- crystal form 1 | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, RR RsiG | | Authors: | Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2021-02-12 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evolution of a sigma-(c-di-GMP)-anti-sigma switch.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LQ3
 
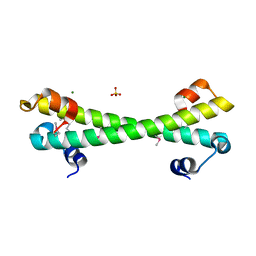 | |
7LQ4
 
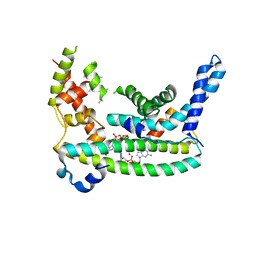 | | Rr (RsiG)2-(c-di-GMP)2-WhiG complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), RsiG, WhiG | | Authors: | Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2021-02-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evolution of a sigma-(c-di-GMP)-anti-sigma switch.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1LIO
 
 | | STRUCTURE OF APO T. GONDII ADENOSINE KINASE | | Descriptor: | adenosine kinase | | Authors: | Schumacher, M.A, Scott, D.M, Mathews, I.I, Ealick, S.E, Brennan, R.G. | | Deposit date: | 2002-04-17 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Toxoplasma gondii adenosine kinase reveal a novel catalytic mechanism and prodrug binding.
J.Mol.Biol., 298, 2000
|
|
1UPF
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V BOUND TO THE DRUG 5-FLUOROURACIL | | Descriptor: | 5-FLUOROURACIL, SULFATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-06-17 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
1UPU
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V, BOUND TO PRODUCT URIDINE-1-MONOPHOSPHATE (UMP) | | Descriptor: | PHOSPHATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-04-16 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
2LIQ
 
 | | Solution structure of CCL2 in complex with glycan | | Descriptor: | CCL2 lectin, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Schubert, M, Bleuler-Martinez, S, Walti, M.A, Egloff, P, Aebi, M, Kuenzler, M, Allain, F.H.-T. | | Deposit date: | 2011-08-30 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
4LSD
 
 | | Myokine structure | | Descriptor: | Fibronectin type III domain-containing protein 5 | | Authors: | Schumacher, M.A, Ohashi, T, Shah, R.S, Chinnam, N, Erickson, H. | | Deposit date: | 2013-07-22 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The structure of irisin reveals a novel intersubunit beta-sheet fibronectin type III (FNIII) dimer: implications for receptor activation.
J.Biol.Chem., 288, 2013
|
|
3OQN
 
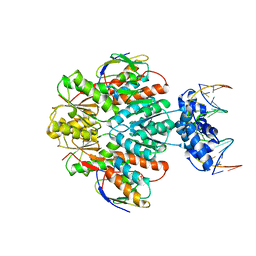 | | Structure of ccpa-hpr-ser46-p-gntr-down cre | | Descriptor: | 5'-D(*AP*TP*GP*GP*TP*AP*CP*CP*GP*CP*TP*TP*TP*CP*AP*A)-3', 5'-D(*TP*TP*GP*AP*AP*AP*GP*CP*GP*GP*TP*AP*CP*CP*AP*T)-3', Catabolite control protein A, ... | | Authors: | Schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
4R25
 
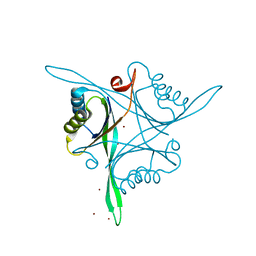 | | Structure of B. subtilis GlnK | | Descriptor: | Nitrogen regulatory PII-like protein, ZINC ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5193 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
4I5B
 
 | |
2RDH
 
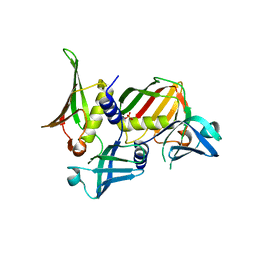 | | Crystal structure of Staphylococcal Superantigen-Like protein 11 | | Descriptor: | PHOSPHATE ION, SODIUM ION, Superantigen-like protein 11 | | Authors: | Chung, M.C, Wines, B.D, Baker, H, Langley, R.J, Baker, E.N, Fraser, J.D. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of staphylococcal superantigen-like protein 11 in complex with sialyl Lewis X reveals the mechanism for cell binding and immune inhibition
Mol.Microbiol., 66, 2007
|
|
3PM1
 
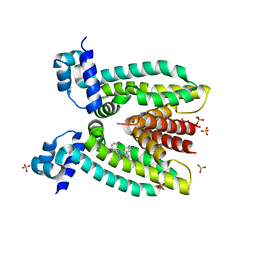 | | Structure of QacR E90Q bound to Ethidium | | Descriptor: | ETHIDIUM, HTH-type transcriptional regulator qacR, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A single acidic residue can guide binding site selection but does not govern QacR cationic-drug affinity.
Plos One, 6, 2011
|
|
3DNT
 
 | | structures of MDT proteins | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein hipA, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
1YM8
 
 | |
2NOQ
 
 | | Structure of ribosome-bound cricket paralysis virus IRES RNA | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S5, ... | | Authors: | Schuler, M, Connell, S.R, Lescoute, A, Giesebrecht, J, Dabrowski, M, Schroeer, B, Mielke, T, Penczek, P.A, Westhof, E, Spahn, C.M.T. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure of the ribosome-bound cricket paralysis virus IRES RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3EZ2
 
 | | Partition protein-ADP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Schumacher, M.A, Dunham, T.D, Xu, W, Funnell, B. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
4KOA
 
 | | Crystal Structure Analysis of 1,5-anhydro-D-fructose reductase from Sinorhizobium meliloti | | Descriptor: | 1,5-anhydro-D-fructose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schu, M, Faust, A, Stosik, B, Kohring, G.-W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2013-05-11 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The structure of substrate-free 1,5-anhydro-D-fructose reductase from Sinorhizobium meliloti 1021 reveals an open enzyme conformation.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2GID
 
 | | Crystal structures of trypanosoma bruciei MRP1/MRP2 | | Descriptor: | mitochondrial RNA-binding protein 1, mitochondrial RNA-binding protein 2 | | Authors: | Schumacher, M.A, Karamooz, E, Zikova, A, Trantirek, L, Lukes, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism.
Cell(Cambridge,Mass.), 126, 2006
|
|
4GCK
 
 | | structure of no-dna complex | | Descriptor: | DNA (5'-D(*GP*TP*GP*AP*GP*TP*AP*CP*TP*CP*AP*C)-3'), Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-07-30 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GFL
 
 | | NO mechanism, slma | | Descriptor: | Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-08-03 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3DNU
 
 | | structure of MDT protein | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Protein hipA | | Authors: | schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3FBR
 
 | | structure of HipA-amppnp-peptide | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Serine/threonine-protein kinase toxin HipA, peptide of EF-Tu | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-11-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
1ZX4
 
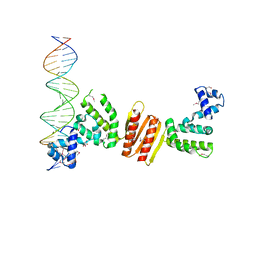 | | Structure of ParB bound to DNA | | Descriptor: | CITRIC ACID, Plasmid Partition par B protein, parS-small DNA centromere site | | Authors: | Schumacher, M.A, Funnell, B.E. | | Deposit date: | 2005-06-06 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structures of ParB bound to DNA reveal mechanism of partition complex formation.
Nature, 438, 2005
|
|
