1OCB
 
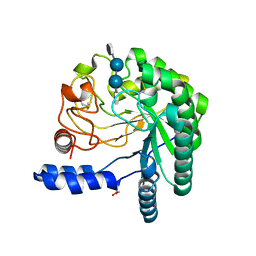 | | Structure of the wild-type cellobiohydrolase Cel6A from Humicolas insolens in complex with a fluorescent substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-amino-4-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-methyl 4-thio-beta-D-glucopyranoside, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
2MFF
 
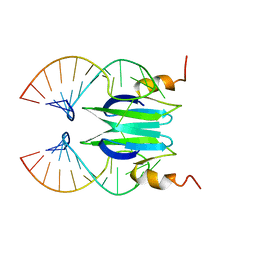 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL3)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL3(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFG
 
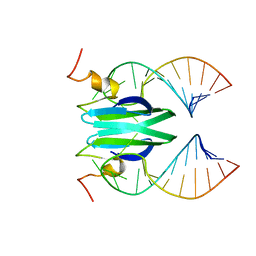 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL4)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL4(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFC
 
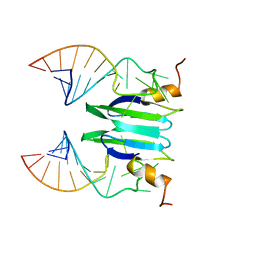 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL1)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL1(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-10 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
1QH6
 
 | | CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE | | Descriptor: | XYLANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Sabini, E, Sulzenbacher, G, Dauter, M, Dauter, Z, Jorgensen, P.L, Schulein, M, Dupont, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase.
Chem.Biol., 6, 1999
|
|
2MFH
 
 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(36-44)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, RsmZ(36-44) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFE
 
 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL2)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL2(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
1OCJ
 
 | | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a THIOPENTASACCHARIDE at 1.3 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1OC5
 
 | | D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with methyl-cellobiosyl-4-deoxy-4-thio-beta-D-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, GLYCEROL, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-06 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1OC7
 
 | | D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with methyl-tetrathio-alpha-d-cellopentoside at 1.1 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-06 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1OC6
 
 | | structure native of the D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS at 1.5 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-06 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1M8M
 
 | | SOLID-STATE MAS NMR STRUCTURE OF THE A-SPECTRIN SH3 DOMAIN | | Descriptor: | SPECTRIN ALPHA CHAIN, BRAIN | | Authors: | Castellani, F, Van Rossum, B, Diehl, A, Schubert, M, Rehbein, K, Oschkinat, H. | | Deposit date: | 2002-07-25 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy
Nature, 420, 2002
|
|
1QHZ
 
 | |
1QI0
 
 | |
2CA3
 
 | | Sulfite dehydrogenase from Starkeya Novella r55m mutant | | Descriptor: | (MOLYBDOPTERIN-S,S)-OXO-MOLYBDENUM, HEME C, SULFATE ION, ... | | Authors: | Bailey, S, Kappler, U, Feng, C, Honeychurch, M.J, Bernhardt, P.V, Tollin, G, Enemark, J.H. | | Deposit date: | 2005-12-16 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for enzymatic sulfite oxidation: how three conserved active site residues shape enzyme activity.
J.Biol.Chem., 284, 2009
|
|
5S4J
 
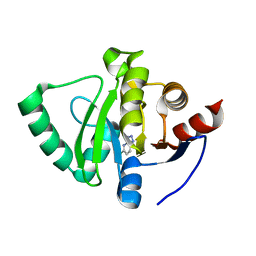 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF054 | | Descriptor: | 6-chlorotetrazolo[1,5-b]pyridazine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4I
 
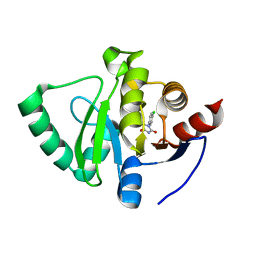 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF051 | | Descriptor: | (5S)-1-(4-chlorophenyl)-5-methylimidazolidine-2,4-dione, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4H
 
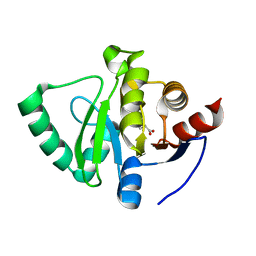 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF048 | | Descriptor: | 1-carbamoylpiperidine-4-carboxylic acid, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.175 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4F
 
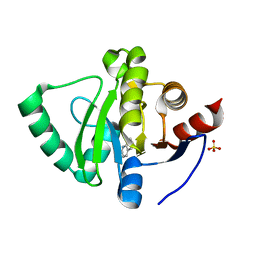 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF003 | | Descriptor: | 1,8-naphthyridine, Non-structural protein 3, SULFATE ION | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4G
 
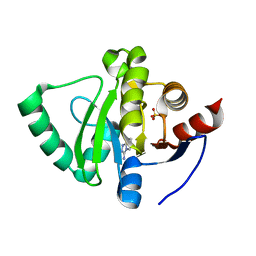 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF005 | | Descriptor: | Non-structural protein 3, SULFATE ION, [1,2,4]triazolo[4,3-a]pyridin-3-amine | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1D7X
 
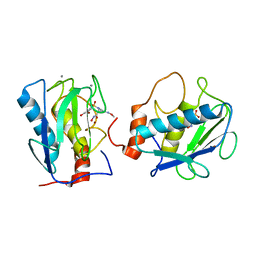 | | CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A MODIFIED PROLINE SCAFFOLD BASED INHIBITOR. | | Descriptor: | CALCIUM ION, N-HYDROXY 1N(4-METHOXYPHENYL)SULFONYL-4-(Z,E-N-METHOXYIMINO)PYRROLIDINE-2R-CARBOXAMIDE, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Cheng, M.Y, Natchus, M.G, De, B, Almstead, N.G, Pikul, S. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and biological evaluation of matrix metalloproteinase inhibitors derived from a modified proline scaffold.
J.Med.Chem., 42, 1999
|
|
1D8M
 
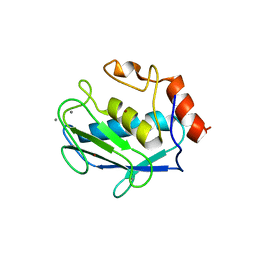 | | CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A HETEROCYCLE-BASED INHIBITOR | | Descriptor: | 1-BENZYL-3-(4-METHOXY-BENZENESULFONYL)-6-OXO-HEXAHYDRO-PYRIMIDINE-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Pikul, S, Dunham, K.M, Almstead, N.G, De, B, Natchus, M.G, Taiwo, Y.O, Williams, L.E, Hynd, B.A, Hsieh, L.C, Janusz, M.J, Gu, F, Mieling, G.E. | | Deposit date: | 1999-10-25 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Heterocycle-based MMP inhibitors with P2' substituents.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
1DYS
 
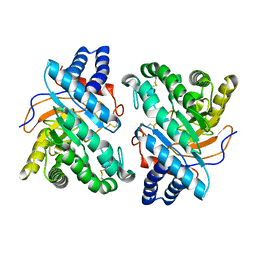 | | Endoglucanase CEL6B from Humicola insolens | | Descriptor: | ENDOGLUCANASE | | Authors: | Davies, G.J, Brzozowski, A.M, Dauter, M, Varrot, A, Schulein, M. | | Deposit date: | 2000-02-08 | | Release date: | 2001-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of Humicola Insolens Family 6 Cellulases: Structure of the Endoglucanase, Cel6B, at 1.6 A Resolution
Biochem.J., 348, 2000
|
|
5S1A
 
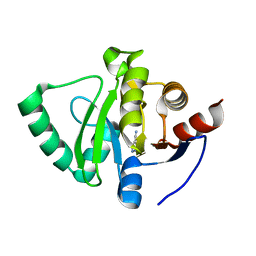 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-43406 | | Descriptor: | 5-amino-3-methyl-1H-pyrazole-4-carbonitrile, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.079 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S1G
 
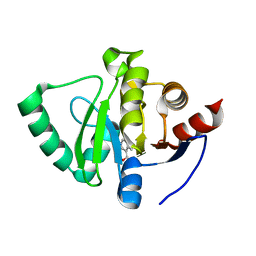 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-108952 | | Descriptor: | (4-methylpyridin-3-yl)methanol, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
