4V68
 
 | | T. thermophilus 70S ribosome in complex with mRNA, tRNAs and EF-Tu.GDP.kirromycin ternary complex, fitted to a 6.4 A Cryo-EM map. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schuette, J.-C, Spahn, C.M.T. | | Deposit date: | 2008-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | GTPase activation of elongation factor EF-Tu by the ribosome during decoding
Embo J., 28, 2009
|
|
2PDZ
 
 | | SOLUTION STRUCTURE OF THE SYNTROPHIN PDZ DOMAIN IN COMPLEX WITH THE PEPTIDE GVKESLV, NMR, 15 STRUCTURES | | Descriptor: | PEPTIDE GVKESLV, SYNTROPHIN | | Authors: | Schultz, J, Hoffmueller, U, Ashurst, J, Krause, G, Schmieder, P, Macias, M, Schneider-Mergener, J, Oschkinat, H. | | Deposit date: | 1997-12-10 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels.
Nat.Struct.Biol., 5, 1998
|
|
6YBQ
 
 | | Engineered glycolyl-CoA carboxylase (quintuple mutant) with bound CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | Authors: | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-28 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
6YBP
 
 | | Propionyl-CoA carboxylase of Methylorubrum extorquens with bound CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | Authors: | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-28 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
6TJV
 
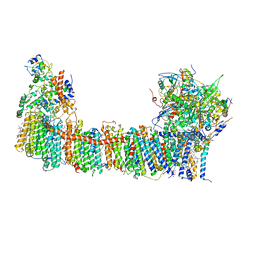 | | Structure of the NDH-1MS complex from Thermosynechococcus elongatus | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, BETA-CAROTENE, ... | | Authors: | Schuller, J.M, Saura, P, Thiemann, J, Schuller, S.K, Gamiz-Hernandez, A.P, Kurisu, G, Nowaczyk, M.M, Kaila, V.R.I. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Redox-coupled proton pumping drives carbon concentration in the photosynthetic complex I.
Nat Commun, 11, 2020
|
|
6HUM
 
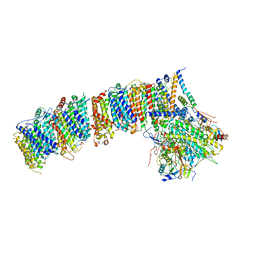 | | Structure of the photosynthetic complex I from Thermosynechococcus elongatus | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, IRON/SULFUR CLUSTER, ... | | Authors: | Schuller, J.M, Schuller, S.K, Kurisu, G, Engel, B.D, Nowaczyk, M.M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer.
Science, 363, 2019
|
|
8F7K
 
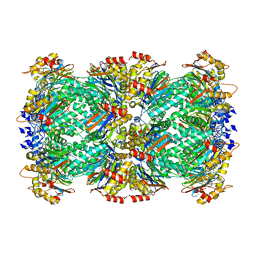 | | Thermoplasma acidophilum 20S proteasome - wild type bound to ZYA | | Descriptor: | N-[(benzyloxy)carbonyl]-L-tyrosyl-D-alanine, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Chuah, J, Smith, D. | | Deposit date: | 2022-11-18 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | High resolution structures define divergent and convergent mechanisms of archaeal proteasome activation.
Commun Biol, 6, 2023
|
|
8F66
 
 | |
8F6A
 
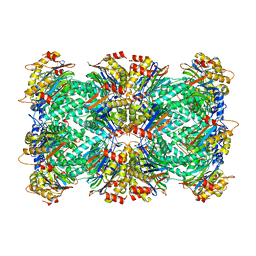 | | Thermoplasma acidophilum 20S proteasome - wild type | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Chuah, J, Smith, D. | | Deposit date: | 2022-11-16 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | High resolution structures define divergent and convergent mechanisms of archaeal proteasome activation.
Commun Biol, 6, 2023
|
|
2CFO
 
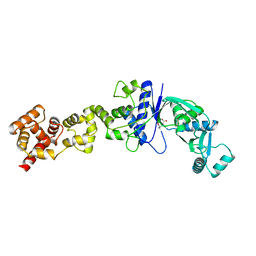 | | Non-Discriminating Glutamyl-tRNA Synthetase from Thermosynechococcus elongatus in Complex with Glu | | Descriptor: | GLUTAMIC ACID, GLUTAMYL-TRNA SYNTHETASE | | Authors: | Schulze, J.O, Nickel, D, Schubert, W.-D, Jahn, D, Heinz, D.W. | | Deposit date: | 2006-02-22 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of a Non-Discriminating Glutamyl- tRNA Synthetase.
J.Mol.Biol., 361, 2006
|
|
2CFB
 
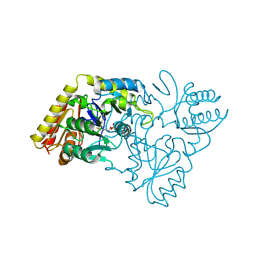 | | Glutamate-1-semialdehyde 2,1-Aminomutase from Thermosynechococcus elongatus | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, GLUTAMATE-1-SEMIALDEHYDE 2,1-AMINOMUTASE | | Authors: | Schulze, J.O, Schubert, W.-D, Moser, J, Jahn, D, Heinz, D.W. | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolutionary Relationship between Initial Enzymes of Tetrapyrrole Biosynthesis
J.Mol.Biol., 358, 2006
|
|
9BUZ
 
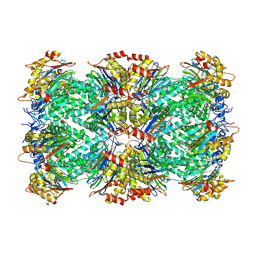 | | Thermoplasma acidophilum 20S proteasome - alphaV24Y | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Chuah, J, Smith, D. | | Deposit date: | 2024-05-18 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Occupancy of the HbYX hydrophobic pocket is sufficient to induce gate opening in the archaeal 20S proteasomes.
Biorxiv, 2024
|
|
6FT6
 
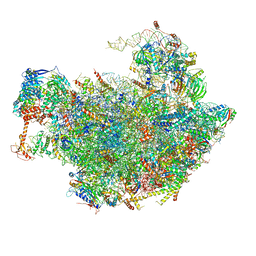 | | Structure of the Nop53 pre-60S particle bound to the exosome nuclear cofactors | | Descriptor: | 25S ribosomal RNA, 5S ribosomal RNA, 60S ribosomal protein L11-A, ... | | Authors: | Schuller, J.M, Falk, S, Conti, E. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the nuclear exosome captured on a maturing preribosome.
Science, 360, 2018
|
|
6FSZ
 
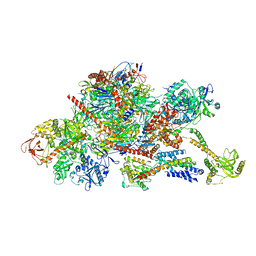 | | Structure of the nuclear RNA exosome | | Descriptor: | ATP-dependent RNA helicase DOB1, Exosome complex component CSL4, Exosome complex component MTR3, ... | | Authors: | Schuller, J.M, Falk, S, Conti, E. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the nuclear exosome captured on a maturing preribosome.
Science, 360, 2018
|
|
6BB4
 
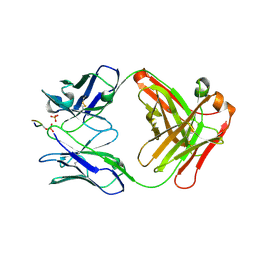 | |
6OEF
 
 | | PolyAla Model of the O-layer from the Type 4 Secretion System of H. pylori | | Descriptor: | PolyAla Model of OMCC O-Layer | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6OEE
 
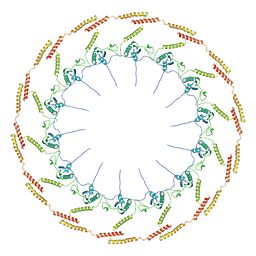 | | Structure of CagT from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagT | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6OEH
 
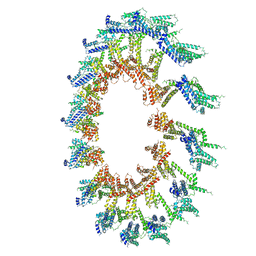 | | PolyAla Model of OMCC I-Layer | | Descriptor: | PolyAla Model of OMCC I-Layer | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6ODI
 
 | | Structure of CagY from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagY | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6ODJ
 
 | | PolyAla Model of the PRC from the Type 4 Secretion System of H. pylori | | Descriptor: | PolyAla Model of PRC from H.pylori | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6OEG
 
 | | Structure of CagX from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagX | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
5TM1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 2,5-bis(2-fluoro-4-hydroxyphenyl)thiophene 1-oxide | | Descriptor: | 2,5-bis(2-fluoro-4-hydroxyphenyl)-1H-1lambda~4~-thiophen-1-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC Analog, (E)-3-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)acrylic acid | | Descriptor: | 3-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}prop-2-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMR
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Cyclofenil-ASC derivative, ethyl (E)-3-(4-(cyclohexylidene(4-hydroxyphenyl)methyl)phenyl)acrylate | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, ethyl 3-{4-[cyclohexylidene(4-hydroxyphenyl)methyl]phenyl}prop-2-enoate | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the estradiol derivative, (8S,9S,13S,14S,E)-17-((4-isopropylphenyl)imino)-13-methyl-7,8,9,11,12,13,14,15,16,17-decahydro-6H-cyclopenta[a]phenanthren-3-ol | | Descriptor: | (9beta,13alpha,17Z)-17-{[4-(propan-2-yl)phenyl]imino}estra-1,3,5(10)-trien-3-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
