6BMB
 
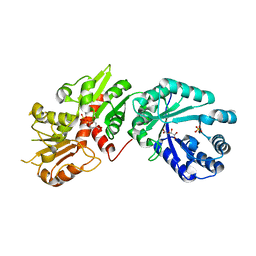 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | 著者 | Christendat, D, Peek, J. | | 登録日 | 2017-11-14 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.077 Å) | | 主引用文献 | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
6BMQ
 
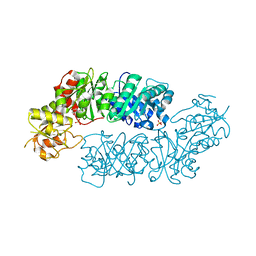 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | 分子名称: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | 著者 | Christendat, D, Peek, J. | | 登録日 | 2017-11-15 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.077 Å) | | 主引用文献 | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
3PWZ
 
 | |
2O7S
 
 | |
2O7Q
 
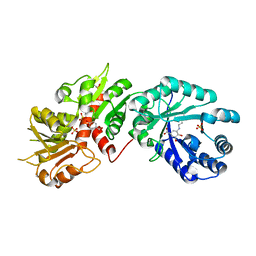 | |
1EJE
 
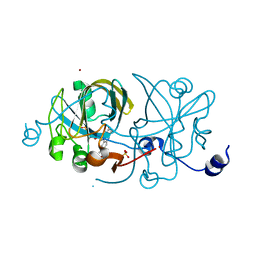 | | CRYSTAL STRUCTURE OF AN FMN-BINDING PROTEIN | | 分子名称: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, NICKEL (II) ION, ... | | 著者 | Christendat, D, Saridakis, V, Bochkarev, A, Arrowsmith, C, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2000-03-02 | | 公開日 | 2000-10-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
1EP0
 
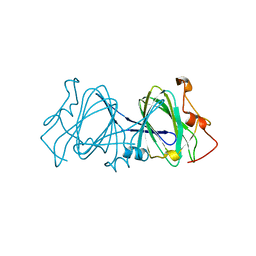 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | 分子名称: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE | | 著者 | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2000-03-24 | | 公開日 | 2000-12-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
1EPZ
 
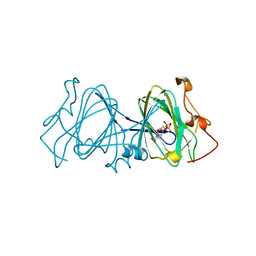 | | CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOASE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH BOUND LIGAND. | | 分子名称: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE, THYMIDINE-5'-DIPHOSPHATE | | 著者 | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C, Edwards, A.M. | | 登録日 | 2000-03-30 | | 公開日 | 2000-12-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
1EIJ
 
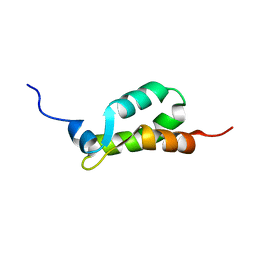 | | NMR ENSEMBLE OF METHANOBACTERIUM THERMOAUTOTROPHICUM PROTEIN 1615 | | 分子名称: | HYPOTHETICAL PROTEIN MTH1615 | | 著者 | Christendat, D, Booth, V, Gernstein, M, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2000-02-25 | | 公開日 | 2000-11-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
1KJN
 
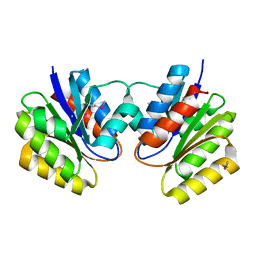 | |
1L1S
 
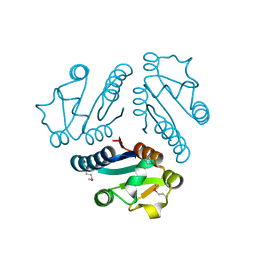 | | Structure of Protein of Unknown Function MTH1491 from Methanobacterium thermoautotrophicum | | 分子名称: | hypothetical protein MTH1491 | | 著者 | Christendat, D, Saridakis, V, Kim, Y, Kumar, P.A, Xu, X, Semesi, A, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-02-19 | | 公開日 | 2002-05-29 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of hypothetical protein MTH1491 from Methanobacterium thermoautotrophicum.
Protein Sci., 11, 2002
|
|
1KUU
 
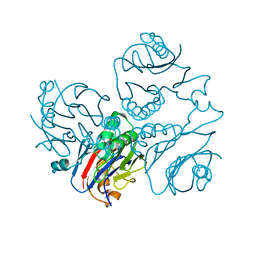 | | CRYSTAL STRUCTURE OF METHANOBACTERIUM THERMOAUTOTROPHICUM CONSERVED PROTEIN MTH1020 REVEALS AN NTN-HYDROLASE FOLD | | 分子名称: | conserved protein | | 著者 | Saridakis, V, Christendat, D, Thygesen, A, Arrowsmith, C.H, Edwards, A.M, Pai, E.F. | | 登録日 | 2002-01-22 | | 公開日 | 2002-05-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF METHANOBACTERIUM THERMOAUTOTROPHICUM CONSERVED PROTEIN MTH1020 REVEALS AN NTN-HYDROLASE FOLD
PROTEINS: STRUCT.,FUNCT.,GENET., 48, 2002
|
|
1K3R
 
 | | Crystal Structure of the Methyltransferase with a Knot from Methanobacterium thermoautotrophicum | | 分子名称: | conserved protein MT0001 | | 著者 | Zarembinski, T.I, Kim, Y, Peterson, K, Christendat, D, Dharamsi, A, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2001-10-03 | | 公開日 | 2002-05-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Deep trefoil knot implicated in RNA binding found in an archaebacterial protein.
Proteins, 50, 2003
|
|
6U60
 
 | | Crystal structure of prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD and L-tyrosine | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Prephenate dehydrogenase, ... | | 著者 | Shabalin, I.G, Hou, J, Kutner, J, Grimshaw, S, Christendat, D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-08-28 | | 公開日 | 2019-09-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
5HMQ
 
 | |
2AS0
 
 | | Crystal Structure of PH1915 (APC 5817): A Hypothetical RNA Methyltransferase | | 分子名称: | hypothetical protein PH1915 | | 著者 | Sun, W, Xu, X, Pavlova, M, Edwards, A.M, Joachimiak, A, Savchenko, A, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-08-22 | | 公開日 | 2005-09-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of a novel SAM-dependent methyltransferase PH1915 from Pyrococcus horikoshii.
Protein Sci., 14, 2005
|
|
3BPX
 
 | | Crystal Structure of MarR | | 分子名称: | 2-HYDROXYBENZOIC ACID, SODIUM ION, Transcriptional regulator | | 著者 | Saridakis, V, Shahinas, D, Xu, X, Christendat, D. | | 登録日 | 2007-12-19 | | 公開日 | 2008-05-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum.
J.Mol.Biol., 377, 2008
|
|
7MQV
 
 | | Crystal structure of truncated (ACT domain removed) prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD | | 分子名称: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Shabalin, I.G, Gritsunov, A, Gabryelska, A, Czub, M.P, Grabowski, M, Cooper, D.R, Christendat, D, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-06 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of Bacillus anthracis prephenate dehydrogenase identified an ACT regulatory domain and a novel mode of metabolic regulation for proteins within the prephenate dehydrogenase family of enzyme
to be published
|
|
3BPV
 
 | | Crystal Structure of MarR | | 分子名称: | Transcriptional regulator | | 著者 | Saridakis, V, Shahinas, D, Xu, X, Christendat, D. | | 登録日 | 2007-12-19 | | 公開日 | 2008-05-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum.
J.Mol.Biol., 377, 2008
|
|
3V85
 
 | |
4OMU
 
 | |
4K28
 
 | | 2.15 Angstrom resolution crystal structure of a shikimate dehydrogenase family protein from Pseudomonas putida KT2440 in complex with NAD+ | | 分子名称: | MANGANESE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | 著者 | Garcia, C, Peek, J, Petit, P, Christendat, D. | | 登録日 | 2013-04-08 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Insights into the function of RifI2: structural and biochemical investigation of a new shikimate dehydrogenase family protein.
Biochim.Biophys.Acta, 1834, 2013
|
|
5TED
 
 | | Effector binding domain of QuiR in complex with shikimate | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, His Tag peptide, Lmo0488 protein | | 著者 | Prezioso, S.M, Christendat, D. | | 登録日 | 2016-09-21 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.889 Å) | | 主引用文献 | Shikimate Induced Transcriptional Activation of Protocatechuate Biosynthesis Genes by QuiR, a LysR-Type Transcriptional Regulator, in Listeria monocytogenes.
J. Mol. Biol., 430, 2018
|
|
5UYY
 
 | | Crystal structure of prephenate dehydrogenase tyrA from Bacillus anthracis in complex with L-tyrosine | | 分子名称: | Prephenate dehydrogenase, TYROSINE | | 著者 | Shabalin, I.G, Hou, J, Cymborowski, M.T, Kwon, K, Christendat, D, Gritsunov, A.O, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-24 | | 公開日 | 2017-03-08 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
5V0S
 
 | | Crystal structure of the ACT domain of prephenate dehydrogenase tyrA from Bacillus anthracis | | 分子名称: | CALCIUM ION, Prephenate dehydrogenase, SULFATE ION | | 著者 | Shabalin, I.G, Hou, J, Cymborowski, M.T, Otwinowski, Z, Kwon, K, Christendat, D, Gritsunov, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-28 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
