6WYA
 
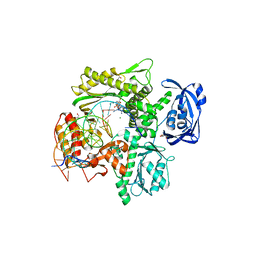 | | RTX (Reverse Transcription Xenopolymerase) in complex with a DNA duplex and dAMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Choi, W.S, He, P, Pothukuchy, A, Gollihar, J, Ellington, A.D, Yang, W. | | Deposit date: | 2020-05-12 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | How a B family DNA polymerase has been evolved to copy RNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WYB
 
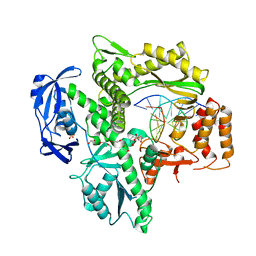 | | RTX (Reverse Transcription Xenopolymerase) in complex with an RNA/DNA hybrid | | Descriptor: | 1,2-ETHANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, DNA polymerase, ... | | Authors: | Choi, W.S, He, P, Pothukuchy, A, Gollihar, J, Ellington, A.D, Yang, W. | | Deposit date: | 2020-05-12 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How a B family DNA polymerase has been evolved to copy RNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7KYQ
 
 | |
7KYS
 
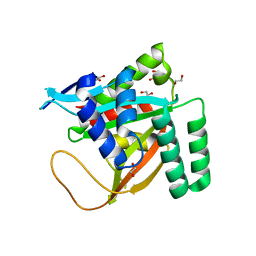 | | Crystal structure of human BCCIP beta (Native2) | | Descriptor: | 1,2-ETHANEDIOL, BRCA2 and CDKN1A-interacting protein, DI(HYDROXYETHYL)ETHER | | Authors: | Choi, W.S, Liu, B, Shen, Z, Yang, W. | | Deposit date: | 2020-12-08 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human BCCIP and implications for binding and modification of partner proteins.
Protein Sci., 30, 2021
|
|
7KQN
 
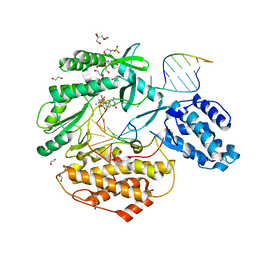 | | Ternary complex of TERT (telomerase reverse transcriptase) with RNA template, DNA primer, an incoming dGTP and a downstream hybrid duplex | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Choi, W.S, Weng, P.J, Yang, W. | | Deposit date: | 2020-11-17 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Flexibility of telomerase in binding the RNA template and DNA telomeric repeat.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7KQM
 
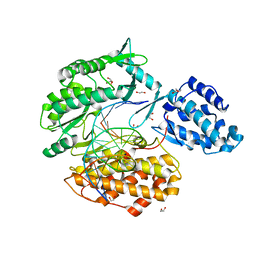 | | Binary complex of TERT (telomerase reverse transcriptase) with RNA/telomeric DNA hybrid | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*TP*TP*CP*TP*TP*TP*GP*TP*GP*CP*AP*CP*CP*TP*G)-3'), ... | | Authors: | Choi, W.S, Weng, P.J, Yang, W. | | Deposit date: | 2020-11-17 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Flexibility of telomerase in binding the RNA template and DNA telomeric repeat.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3E57
 
 | |
3NIH
 
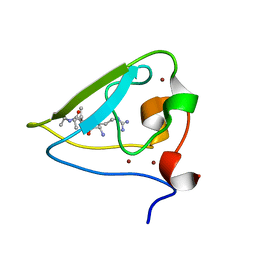 | | The structure of UBR box (RIAAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIL
 
 | | The structure of UBR box (RDAA) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIK
 
 | | The structure of UBR box (REAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide REAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIS
 
 | | The structure of UBR box (native2) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
7LBX
 
 | |
7LBW
 
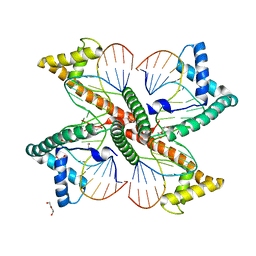 | |
4CAK
 
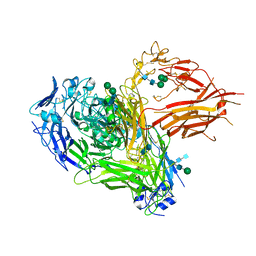 | | Three-dimensional reconstruction of intact human integrin alphaIIbbeta3 in a phospholipid bilayer nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-IIb, ... | | Authors: | Choi, W.S, Rice, W.J, Stokes, D.L, Coller, B.S. | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (20.5 Å) | | Cite: | Three-Dimensional Reconstruction of Intact Human Integrin Alphaiibbeta3; New Implications for Activation-Dependent Ligand Binding.
Blood, 122, 2013
|
|
3NIJ
 
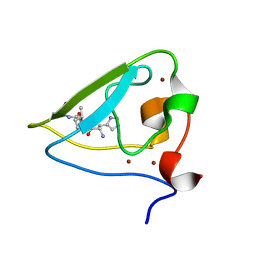 | | The structure of UBR box (HIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide HIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIN
 
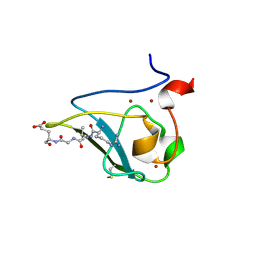 | | The structure of UBR box (RLGES) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RLGES, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIT
 
 | | The structure of UBR box (native1) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NII
 
 | | The structure of UBR box (KIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide KIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIM
 
 | | The structure of UBR box (RRAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RRAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
6CUD
 
 | | Structure of the human TRPC3 in a lipid-occupied, closed state | | Descriptor: | (2R)-3-hydroxypropane-1,2-diyl dihexanoate, (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lu, W, Du, J, Fan, C, Choi, W. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human lipid-gated cation channel TRPC3.
Elife, 7, 2018
|
|
6UIW
 
 | |
6UIV
 
 | |
6UIX
 
 | | Cryo-EM structure of human CALHM2 gap junction | | Descriptor: | Calcium homeostasis modulator protein 2 | | Authors: | Lu, W, Du, J, Choi, W. | | Deposit date: | 2019-10-01 | | Release date: | 2019-11-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structures and gating mechanism of human calcium homeostasis modulator 2.
Nature, 576, 2019
|
|
6KRT
 
 | | monodehydroascorbate reductase, MDHAR, from Antarctic hairgrass Deschampsia antarctica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, monodehydroascorbate reductase | | Authors: | Park, A.K, Do, H, Lee, J.H, Kim, H, Choi, W, Kim, I.S, Kim, H.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | monodehydroascorbate reductase, MDHAR, from Antarctic hairgrass Deschampsia antarctica
To Be Published
|
|
4FZV
 
 | | Crystal structure of the human MTERF4:NSUN4:SAM ternary complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative methyltransferase NSUN4, ... | | Authors: | Guja, K.E, Yakubovskaya, E, Mejia, E, Castano, S, Hambardjieva, E, Choi, W.S, Garcia-Diaz, M. | | Deposit date: | 2012-07-08 | | Release date: | 2012-10-03 | | Last modified: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.9996 Å) | | Cite: | Structure of the Essential MTERF4:NSUN4 Protein Complex Reveals How an MTERF Protein Collaborates to Facilitate rRNA Modification.
Structure, 20, 2012
|
|
