4GMX
 
 | |
5J3V
 
 | |
4FDD
 
 | | Crystal structure of KAP beta2-PY-NLS | | Descriptor: | RNA-binding protein FUS, Transportin-1 | | Authors: | Zhang, Z.C, Chook, Y.M. | | Deposit date: | 2012-05-28 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS).
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HB4
 
 | |
4HAZ
 
 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(R543S,K548E,K579Q)-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HAW
 
 | |
4HAY
 
 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(K548E,K579Q)-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HB3
 
 | |
4HB0
 
 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(K541Q,K542Q,R543S,K545Q,K548Q,K579Q)-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HAX
 
 | |
4HB2
 
 | | Crystal structure of CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HAT
 
 | |
4HAU
 
 | |
4HAV
 
 | |
6CIT
 
 | |
2M4M
 
 | | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata | | Descriptor: | hypothetical protein | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Patel, H, Seidel, R.D, Chook, Y.M, Rout, M.P, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata
To be Published
|
|
7CB7
 
 | | 1.7A resolution structure of SARS-CoV-2 main protease (Mpro) in complex with broad-spectrum coronavirus protease inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y, Hung, M.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
5JLJ
 
 | | Crystal Structure of KPT8602 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, CHLORIDE ION, Exportin-1, ... | | Authors: | Fung, H.Y, Chook, Y.M. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies.
Leukemia, 30, 2016
|
|
2BD0
 
 | | Chlorobium tepidum Sepiapterin Reductase complexed with NADP and Sepiapterin | | Descriptor: | BIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, sepiapterin reductase | | Authors: | Supangat, S, Seo, K.H, Choi, Y.K, Park, Y.S, Lee, K.H. | | Deposit date: | 2005-10-19 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Chlorobium tepidum sepiapterin reductase complex reveals the novel substrate binding mode for stereospecific production of L-threo-tetrahydrobiopterin
J.Biol.Chem., 281, 2006
|
|
7CAM
 
 | | SARS-CoV-2 main protease (Mpro) apo structure (space group P212121) | | Descriptor: | 3C-like proteinase | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y. | | Deposit date: | 2020-06-09 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
1Z8G
 
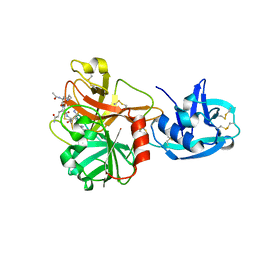 | | Crystal structure of the extracellular region of the transmembrane serine protease hepsin with covalently bound preferred substrate. | | Descriptor: | ACE-LYS-GLN-LEU-ARG-Chloromethylketone, Serine protease hepsin | | Authors: | Herter, S, Piper, D.E, Aaron, W, Gabriele, T, Cutler, G, Cao, P, Bhatt, A.S, Choe, Y, Craik, C.S, Walker, N, Meininger, D, Hoey, T, Austin, R.J. | | Deposit date: | 2005-03-30 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membrane-anchored serine protease implicated in prostate and ovarian cancers
Biochem.J., 390, 2005
|
|
1Q90
 
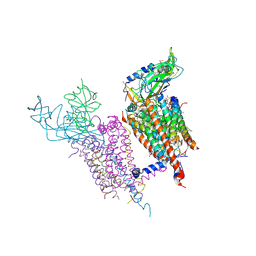 | | Structure of the cytochrome b6f (plastohydroquinone : plastocyanin oxidoreductase) from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 8-HYDROXY-5,7-DIMETHOXY-3-METHYL-2-TRIDECYL-4H-CHROMEN-4-ONE, ... | | Authors: | Stroebel, D, Choquet, Y, Popot, J.-L, Picot, D. | | Deposit date: | 2003-08-22 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An Atypical Haem in the Cytochrome B6F Complex
Nature, 426, 2003
|
|
5DI9
 
 | |
5UWW
 
 | |
5UWJ
 
 | |
