4X04
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM CITROBACTER KOSERI (CKO_04899, TARGET EFI-510094) WITH BOUND D-glucuronate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, beta-D-glucopyranuronic acid, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM CITROBACTER KOSERI (CKO_04899, TARGET EFI-510094) WITH BOUND D-glucuronate
To be published
|
|
6SM1
 
 | | Wild type immunoglobulin light chain (WT-1) | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Immunoglobulin lambda variable 2-14, ... | | Authors: | Kazman, P, Vielberg, M.-T, Cendales, M.D.P, Hunziger, L, Weber, B, Hegenbart, U, Zacharias, M, Koehler, R, Schoenland, S, Groll, M, Buchner, J. | | Deposit date: | 2019-08-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fatal amyloid formation in a patient's antibody light chain is caused by a single point mutation.
Elife, 9, 2020
|
|
4XF5
 
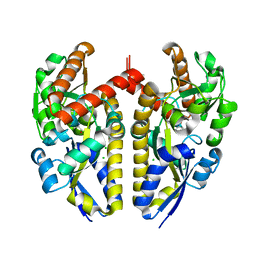 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound (S)-(+)-2-Amino-1-propanol. | | Descriptor: | (2S)-2-aminopropan-1-ol, CHLORIDE ION, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-26 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound (S)-(+)-2-Amino-1-propanol.
To be published
|
|
1XDT
 
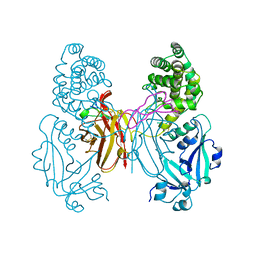 | | COMPLEX OF DIPHTHERIA TOXIN AND HEPARIN-BINDING EPIDERMAL GROWTH FACTOR | | Descriptor: | DIPHTHERIA TOXIN, HEPARIN-BINDING EPIDERMAL GROWTH FACTOR | | Authors: | Louie, G.V, Yang, W, Bowman, M.E, Choe, S. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex of diphtheria toxin with an extracellular fragment of its receptor.
Mol.Cell, 1, 1997
|
|
6JJA
 
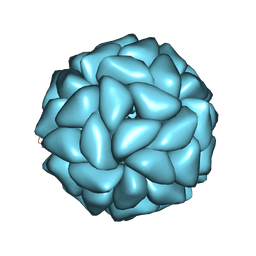 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii extra small virus (XSV) VLP | | Descriptor: | CALCIUM ION, Nucleocapsid protein CP17 | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii extra small virus (XSV) VLP
To Be Published
|
|
1B92
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-19 | | Release date: | 1999-07-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1B9D
 
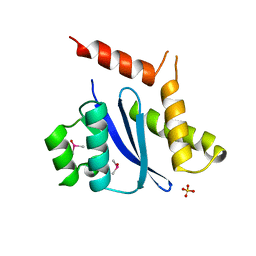 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
7BDV
 
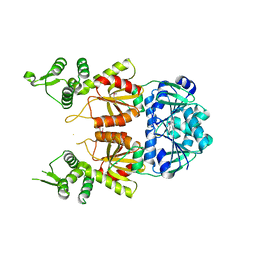 | | Structure of Can2 from Sulfobacillus thermosulfidooxidans in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Can2, Cyclic tetraadenosine monophosphate (cA4) | | Authors: | McQuarrie, S, McMahon, S.A, Gloster, T.M, White, M.F, Graham, S, Zhu, W, Gruschow, S. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The CRISPR ancillary effector Can2 is a dual-specificity nuclease potentiating type III CRISPR defence.
Nucleic Acids Res., 49, 2021
|
|
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
1XUE
 
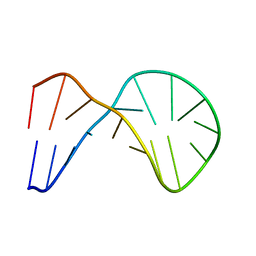 | |
4N0V
 
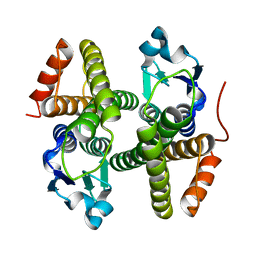 | | Crystal structure of a glutathione S-transferase domain-containing protein (Marinobacter aquaeolei VT8), Target EFI-507332 | | Descriptor: | Glutathione S-transferase, N-terminal domain | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-02 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a glutathione S-transferase domain-containing protein (Marinobacter aquaeolei VT8), Target EFI-507332
TO BE PUBLISHED
|
|
1XS3
 
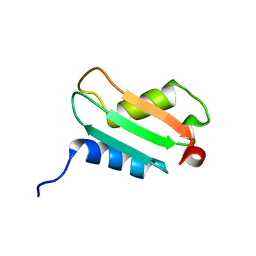 | | Solution Structure Analysis of the XC975 protein | | Descriptor: | hypothetical protein XC975 | | Authors: | Chin, K.-H, Lin, F.-Y, Hu, Y.-C, Sze, K.-H, Lyu, P.-C, Chou, S.-H. | | Deposit date: | 2004-10-18 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Letter to the Editor: NMR structure note - Solution structure of a bacterial BolA-like protein XC975 from a plant pathogen Xanthomonas campestris pv. campestris
J.Biomol.Nmr, 31, 2005
|
|
1B9F
 
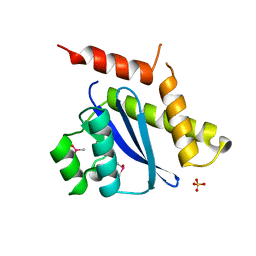 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
5U6Y
 
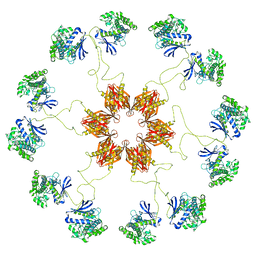 | | Pseudo-atomic model of the CaMKIIa holoenzyme. | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Myers, J, Reichow, S.L. | | Deposit date: | 2016-12-09 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | The CaMKII holoenzyme structure in activation-competent conformations.
Nat Commun, 8, 2017
|
|
1J3I
 
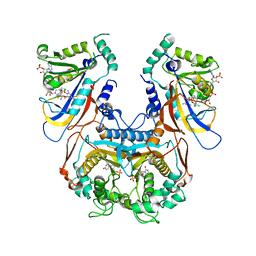 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with WR99210, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 6,6-DIMETHYL-1-[3-(2,4,5-TRICHLOROPHENOXY)PROPOXY]-1,6-DIHYDRO-1,3,5-TRIAZINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Yuvaniyama, J, Chitnumsub, P, Kamchonwongpaisan, S, Vanichtanankul, J, Sirawaraporn, W, Taylor, P, Walkinshaw, M, Yuthavong, Y. | | Deposit date: | 2003-02-03 | | Release date: | 2003-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Insights into antifolate resistance from malarial DHFR-TS structures.
NAT.STRUCT.BIOL., 10, 2003
|
|
6JJC
 
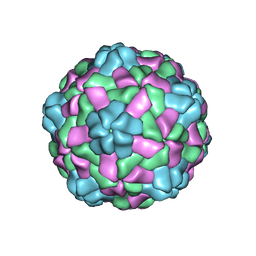 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP
To Be Published
|
|
6JJD
 
 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) full VLP | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) full VLP
To Be Published
|
|
6QF1
 
 | | X-Ray structure of Proteinase K crystallized on a silicon chip | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Proteinase K | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6QF2
 
 | | X-Ray structure of Thermolysin crystallized on a silicon chip | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CALCIUM ION, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
8DM3
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with Fab 4A8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 4A8 heavy chain, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM2
 
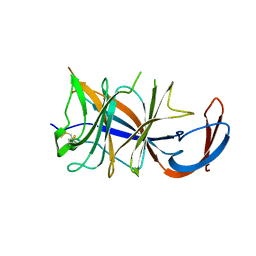 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein (focused refinement of NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM1
 
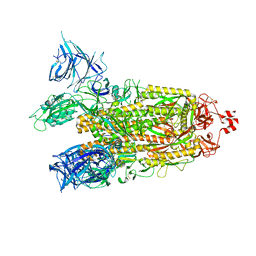 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM4
 
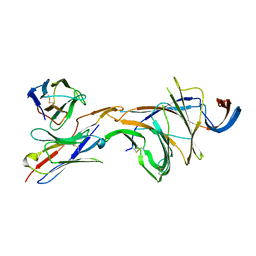 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with Fab 4A8 (focused refinement of NTD and 4A8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 4A8 heavy chain, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM6
 
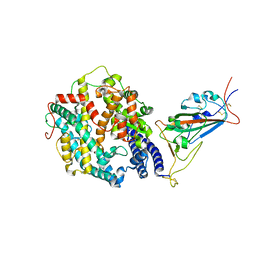 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DMA
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
