6QF3
 
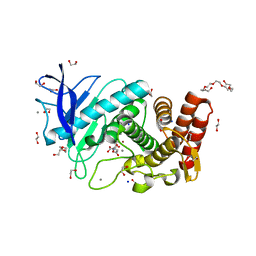 | | X-Ray structure of Thermolysin soaked with sodium aspartate on a silicon chip | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ASPARTIC ACID, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
1U4F
 
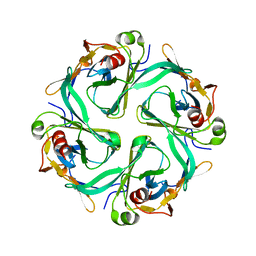 | | Crystal Structure of Cytoplasmic Domains of IRK1 (Kir2.1) channel | | Descriptor: | Inward rectifier potassium channel 2 | | Authors: | Pegan, S, Arrabit, C, Zhou, W, Kwiatkowski, W, Slesinger, P.A, Choe, S. | | Deposit date: | 2004-07-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification
Nat.Neurosci., 8, 2005
|
|
6QF4
 
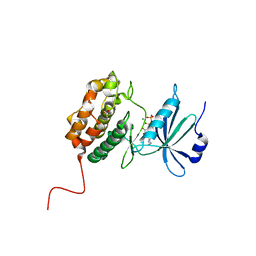 | | X-Ray structure of human Serine/Threonine Kinase 17B (STK17B) aka DRAK2 in complex with ADP obtained by on-chip soaking | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6QF5
 
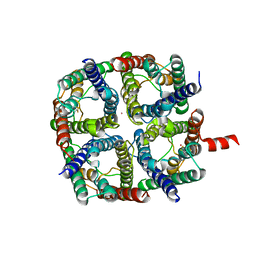 | | X-Ray structure of human Aquaporin 2 crystallized on a silicon chip | | Descriptor: | Aquaporin-2, CADMIUM ION | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
5V7V
 
 | | Cryo-EM structure of ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ERAD-associated E3 ubiquitin-protein ligase component HRD3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mi, W, Schoebel, S, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
5U6Y
 
 | | Pseudo-atomic model of the CaMKIIa holoenzyme. | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Myers, J, Reichow, S.L. | | Deposit date: | 2016-12-09 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | The CaMKII holoenzyme structure in activation-competent conformations.
Nat Commun, 8, 2017
|
|
3UJ2
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE FROM ANAEROSTIPES CACCAE (EFI TARGET EFI-502054) WITH BOUND MG AND SULFATE | | Descriptor: | Enolase 1, MAGNESIUM ION, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE FROM ANAEROSTIPES CACCAE (EFI TARGET EFI-502054) WITH BOUND MG AND SULFATE
to be published
|
|
3UAP
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501774) from methylococcus capsulatus str. bath | | Descriptor: | GLYCEROL, Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Methylococcus Capsulatus
To be Published
|
|
3UBK
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501770) from leptospira interrogans | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutathione transferase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Leptospira Interrogans
To be Published
|
|
1XUE
 
 | |
1ZKZ
 
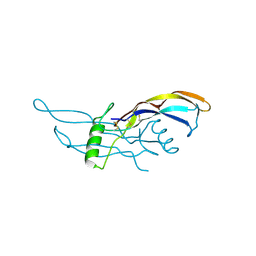 | | Crystal Structure of BMP9 | | Descriptor: | Growth/differentiation factor 2 | | Authors: | Brown, M.A, Zhao, Q, Baker, K.A, Naik, C, Chen, C, Pukac, L, Singh, M, Tsareva, T, Parice, Y, Mahoney, A, Roschke, V, Sanyal, I, Choe, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of BMP-9 and functional interactions with pro-region and receptors
J.Biol.Chem., 280, 2005
|
|
3TOY
 
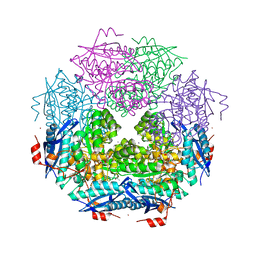 | | CRYSTAL STRUCTURE OF ENOLASE BRADO_4202 (TARGET EFI-501651) FROM Bradyrhizobium sp. ORS278 WITH CALCIUM AND ACETATE BOUND | | Descriptor: | ACETATE ION, CALCIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammond, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF MANDELATE RACEMASE FROM Bradyrhizobium sp. ORS278
To be Published
|
|
3TWB
 
 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 complexed with magnesium and gluconic acid | | Descriptor: | CHLORIDE ION, D-gluconic acid, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
2L2Z
 
 | | Thiostrepton, reduced at N-CA bond of residue 14 | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-06-26 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2L2W
 
 | | Thiostrepton | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2L2Y
 
 | | Thiostrepton, epimer form of residue 9 | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-06-26 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2L0I
 
 | | Solution structure of Rtt103 CTD-interacting domain bound to a Ser2 phosphorylated CTD peptide | | Descriptor: | DNA-directed RNA polymerase, Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S.L, Kim, M, Suh, H, Leeper, T.C, Yang, F, Mutschler, H, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2010-07-06 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3TTE
 
 | | Crystal structure of enolase brado_4202 (target EFI-501651) from Bradyrhizobium complexed with magnesium and mandelic acid | | Descriptor: | (S)-MANDELIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammond, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-14 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mandelate Racemase from Bradyrhizobium Sp. Ors278
To be Published
|
|
3TW9
 
 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative dehydratase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
3UBL
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501770) from leptospira interrogans with gsh bound | | Descriptor: | CHLORIDE ION, GLUTATHIONE, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Leptospira Interrogans
To be Published
|
|
2KM4
 
 | | Solution structure of Rtt103 CTD interacting domain | | Descriptor: | Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S, Kim, M, Leeper, T.C, Becker, R, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2009-07-20 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2N34
 
 | | NMR assignments and solution structure of the JAK interaction region of SOCS5 | | Descriptor: | Suppressor of cytokine signaling 5 | | Authors: | Chandrashekaran, I.R, Mohanty, B, Linossi, E.M, Nicholson, S.E, Babon, J, Norton, R.S, Dagley, L.F, Leung, E.W.W, Murphy, J.M. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Functional Characterization of the Conserved JAK Interaction Region in the Intrinsically Disordered N-Terminus of SOCS5.
Biochemistry, 54, 2015
|
|
2F9W
 
 | | Structure of the type III CoaA from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PANTOTHENOIC ACID, ... | | Authors: | Leonardi, R, Yun, M.K, Chohnan, S, White, S.W, Rock, C.O, Jackowski, S. | | Deposit date: | 2005-12-06 | | Release date: | 2006-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prokaryotic Type II and Type III Pantothenate Kinases: The Same Monomer Fold Creates Dimers with Distinct Catalytic Properties.
Structure, 14, 2006
|
|
3V3W
 
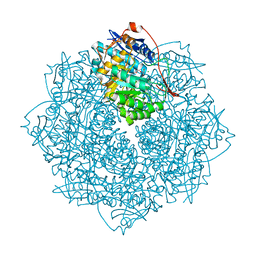 | | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and glycerol | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and glycerol
to be published
|
|
3V4B
 
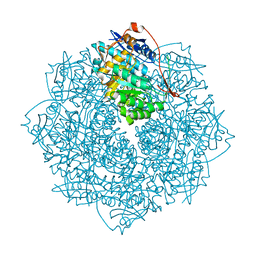 | | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and L-tartrate | | Descriptor: | CHLORIDE ION, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase from the soil bacterium Cellvibrio japonicus (TARGET EFI-502161) with bound MG and l-tartrate
to be published
|
|
