3ARA
 
 | | Discovery of Novel Uracil Derivatives as Potent Human dUTPase Inhibitors | | Descriptor: | 1-[3-({(2R)-2-[hydroxy(diphenyl)methyl]pyrrolidin-1-yl}sulfonyl)propyl]pyrimidine-2,4(1H,3H)-dione, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Chong, K.T, Miyakoshi, H, Miyahara, S, Fukuoka, M. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and discovery of N-carbonylpyrrolidine- or N-sulfonylpyrrolidine-containing uracil derivatives as potent human deoxyuridine triphosphatase inhibitors
J.Med.Chem., 55, 2012
|
|
2ADV
 
 | | Crystal Structures Of Glutaryl 7-Aminocephalosporanic Acid Acylase: mutational study of activation mechanism | | Descriptor: | Glutaryl 7- Aminocephalosporanic Acid Acylase | | Authors: | Kim, J.K, Yang, I.S, Shin, H.J, Cho, K.J, Ryu, E.K, Kim, S.H, Park, S.S, Kim, K.H. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2AE4
 
 | | Glutaryl 7-Aminocephalosporanic Acid Acylase: mutational study of activation mechanism | | Descriptor: | GLYCEROL, Glutaryl 7-Aminocephalosporanic Acid Acylase, SULFATE ION | | Authors: | Kim, J.K, Yang, I.S, Shin, H.J, Cho, K.J, Ryu, E.K, Kim, S.H, Park, S.S, Kim, K.H. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2AE3
 
 | | Glutaryl 7-Aminocephalosporanic Acid Acylase: mutational study of activation mechanism | | Descriptor: | GLYCEROL, Glutaryl 7-Aminocephalosporanic Acid Acylase | | Authors: | Kim, J.K, Yang, I.S, Shin, H.J, Cho, K.J, Ryu, E.K, Kim, S.H, Park, S.S, Kim, K.H. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3BVK
 
 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
3BVL
 
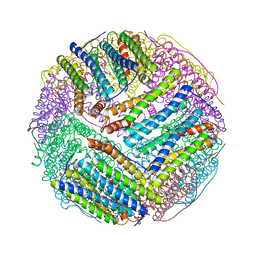 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
3BVF
 
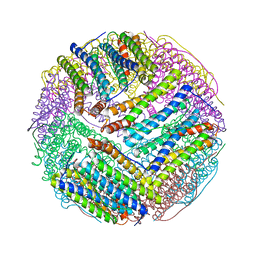 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL, ... | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
3BVI
 
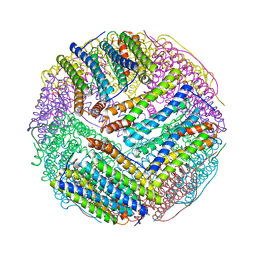 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
2AE5
 
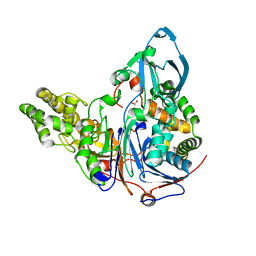 | | Glutaryl 7-Aminocephalosporanic Acid Acylase: mutational study of activation mechanism | | Descriptor: | GLYCEROL, Glutaryl 7-Aminocephalosporanic Acid Acylase, SULFATE ION | | Authors: | Kim, J.K, Yang, I.S, Shin, H.J, Cho, K.J, Ryu, E.K, Kim, S.H, Park, S.S, Kim, K.H. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Insight into autoproteolytic activation from the structure of cephalosporin acylase: a protein with two proteolytic chemistries.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1OPY
 
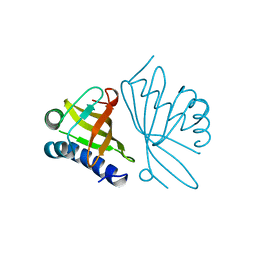 | | KSI | | Descriptor: | DELTA5-3-KETOSTEROID IOSMERASE | | Authors: | Kim, S.-W, Cha, S.-S, Cho, H.-S, Kim, J.-S, Ha, N.-C, Cho, M.-J, Choi, K.-Y, Oh, B.-H. | | Deposit date: | 1997-05-23 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures of delta5-3-ketosteroid isomerase with and without a reaction intermediate analogue.
Biochemistry, 36, 1997
|
|
1EFE
 
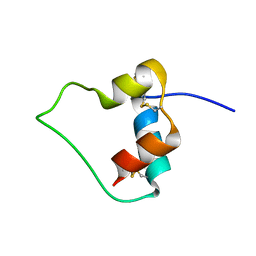 | | AN ACTIVE MINI-PROINSULIN, M2PI | | Descriptor: | MINI-PROINSULIN | | Authors: | Cho, Y, Chang, S.G, Choi, K.D, Shin, H, Ahn, B, Kim, K.S. | | Deposit date: | 2000-02-08 | | Release date: | 2000-03-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Active Mini-Proinsulin, M2PI: Inter-chain Flexibility is Crucial for Insulin Activity
J.Biochem.Mol.Biol., 33, 2000
|
|
1HDU
 
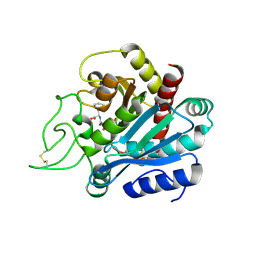 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with aminocarbonylphenylalanine at 1.75 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(AMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1HEE
 
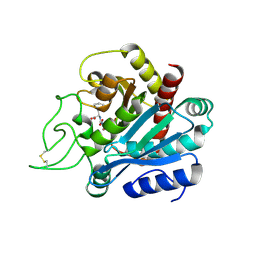 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with L-N-hydroxyaminocarbonyl phenylalanine at 2.3 A | | Descriptor: | CARBOXYPEPTIDASE A, L-[(N-HYDROXYAMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-22 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1HDQ
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with D-N-hydroxyaminocarbonyl phenylalanine at 2.3 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(N-HYDROXYAMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
4LVH
 
 | | Insight into highly conserved H1 subtype-specific epitopes in influenza virus hemagglutinin | | Descriptor: | CALCIUM ION, Hemagglutinin, MONOCLONAL ANTIBODY H-CHAIN, ... | | Authors: | Kim, K.H, Cho, K.J, Kim, S, Seok, J.H, Lee, J.-H. | | Deposit date: | 2013-07-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insight into highly conserved h1 subtype-specific epitopes in influenza virus hemagglutinin
Plos One, 9, 2014
|
|
2MXD
 
 | | Solution structure of VPg of porcine sapovirus | | Descriptor: | Viral protein genome-linked | | Authors: | Kim, J, Hwang, H, Min, H, Yun, H, Cho, K, Pelton, J.G, Wemmer, D.E, Lee, C. | | Deposit date: | 2014-12-24 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the porcine sapovirus VPg core reveals a stable three-helical bundle with a conserved surface patch.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
3EGM
 
 | | Structural basis of iron transport gating in Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Shin, H.J, Lee, J.H. | | Deposit date: | 2008-09-11 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of ferritin from Helicobacter pylori reveals unusual conformational changes for iron uptake.
J.Mol.Biol., 390, 2009
|
|
3BVE
 
 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
4EDB
 
 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
4F15
 
 | | Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-05-06 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses
To be Published
|
|
4EDA
 
 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
4Q6P
 
 | | Structural analysis of the Zn-form I of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6Q
 
 | | Structural analysis of the Zn-form II of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6N
 
 | | Structural analysis of the tripeptide-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6M
 
 | | Structural analysis of the apo-form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
