1TXM
 
 | | SCORPION TOXIN (MAUROTOXIN) FROM SCORPIO MAURUS, NMR, 35 STRUCTURES | | Descriptor: | MAUROTOXIN | | Authors: | Blanc, E, Sabatier, J.-M, Kharrat, R, Meunier, S, El Ayeb, M, Van Rietschoten, J, Darbon, H. | | Deposit date: | 1996-11-19 | | Release date: | 1997-06-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of maurotoxin, a scorpion toxin from Scorpio maurus, with high affinity for voltage-gated potassium channels.
Proteins, 29, 1997
|
|
3TIO
 
 | | Crystal structures of yrdA from Escherichia coli, a homologous protein of gamma-class carbonic anhydrase, show possible allosteric conformations | | Descriptor: | PHOSPHATE ION, Protein YrdA, ZINC ION | | Authors: | Park, H.M, Choi, J.W, Lee, J.E, Jung, C.H, Kim, B.Y, Kim, J.S. | | Deposit date: | 2011-08-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structures of the gamma-class carbonic anhydrase homologue YrdA suggest a possible allosteric switch
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8ZU6
 
 | |
7KD9
 
 | | Crystal Structure of Gallic Acid Decarboxylase from Arxula adeninivorans | | Descriptor: | Gallate decarboxylase, POTASSIUM ION | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
1XDK
 
 | | Crystal Structure of the RARbeta/RXRalpha Ligand Binding Domain Heterodimer in Complex with 9-cis Retinoic Acid and a Fragment of the TRAP220 Coactivator | | Descriptor: | (9cis)-retinoic acid, Retinoic acid receptor RXR-alpha, Retinoic acid receptor, ... | | Authors: | Pogenberg, V, Guichou, J.F, Vivat-Hannah, V, Kammerer, S, Perez, E, Germain, P, De Lera, A.R, Gronemeyer, H, Royer, C.A, Bourguet, W. | | Deposit date: | 2004-09-07 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CHARACTERIZATION OF THE INTERACTION BETWEEN RAR/RXR HETERODIMERS AND TRANSCRIPTIONAL COACTIVATORS THROUGH STRUCTURAL AND FLUORESCENCE ANISOTROPY STUDIES
J.Biol.Chem., 280, 2005
|
|
7JI1
 
 | |
6XLQ
 
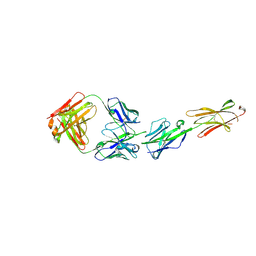 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the CTX-2026 Fab | | Descriptor: | Butyrophilin subfamily 3 member A1, CTX-2026 Heavy Chain, CTX-2026 Light Chain | | Authors: | Payne, K.K, Mine, J.A, Biswas, S, Chaurio, R.A, Perales-Puchalt, A, Anadon, C.M, Costich, T.L, Harro, C.M, Walrath, J, Ming, Q, Tcyganov, E, Buras, A.L, Rigolizzo, K.E, Mandal, G, Lajoie, J, Ophir, M, Tchou, J, Marchion, D, Luca, V.C, Bobrowicz, P, McLaughlin, B, Eskiocak, U, Schmidt, M, Cubillos-Ruiz, J.R, Rodriguez, P.C, Gabrilovich, D.I, Conejo-Garcia, J.R. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BTN3A1 governs antitumor responses by coordinating alpha beta and gamma delta T cells.
Science, 369, 2020
|
|
7JMR
 
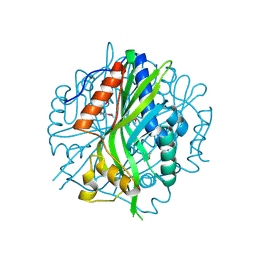 | | Crystal structure of the pea pathogenicity protein 2 from Madurella mycetomatis | | Descriptor: | CALCIUM ION, POTASSIUM ION, Pea pathogenicity protein 2 | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
7JMV
 
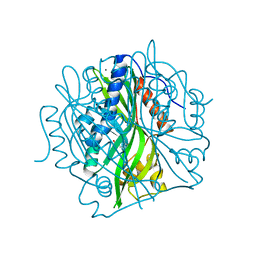 | | Crystal structure of the pea pathogenicity protein 2 from Madurella mycetomatis complexed with 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-08-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
6FJ3
 
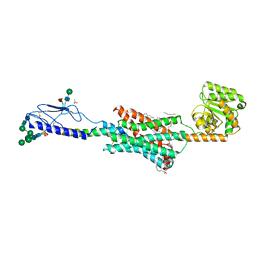 | | High resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist. | | Descriptor: | ACETIC ACID, CHLORIDE ION, OLEIC ACID, ... | | Authors: | Ehrenmann, J, Schoppe, J, Klenk, C, Rappas, M, Kummer, L, Dore, A.S, Pluckthun, A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7LOH
 
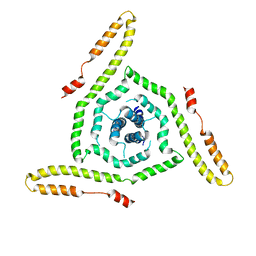 | | Structure of the HIV-1 gp41 transmembrane domain and cytoplasmic tail | | Descriptor: | Transmembrane protein gp41 | | Authors: | Piai, A, Fu, Q, Sharp, A.K, Bighi, B, Brown, A.M, Chou, J.J. | | Deposit date: | 2021-02-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Model of the Entire Membrane-Interacting Region of the HIV-1 Fusion Protein and Its Perturbation of Membrane Morphology.
J.Am.Chem.Soc., 143, 2021
|
|
1Y4Y
 
 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
4EEY
 
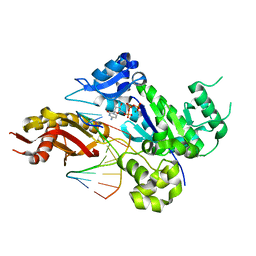 | | Crystal structure of human DNA polymerase eta in ternary complex with a cisplatin DNA adduct | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*TP*GP*GP*TP*CP*TP*CP*CP*TP*CP*C)-3', 5'-D(*TP*GP*GP*AP*GP*GP*AP*GP*A)-3', ... | | Authors: | Ummat, A, Rechkoblit, O, Jain, R, Choudhury, J.R, Johnson, R.E, Silverstein, T.D, Buku, A, Lone, S, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for cisplatin DNA damage tolerance by human polymerase {eta} during cancer chemotherapy.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1Y50
 
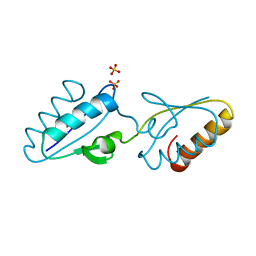 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) F29W mutant domain_swapped dimer | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
7K7A
 
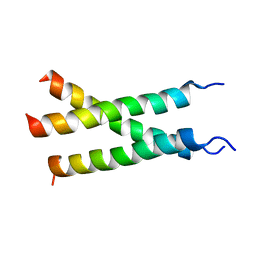 | | Transmembrane structure of TNFR1 | | Descriptor: | Tumor necrosis factor receptor superfamily member 1A | | Authors: | Zhao, L, Chou, J. | | Deposit date: | 2020-09-22 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Diversity and Similarity of Transmembrane Trimerization of TNF Receptors.
Front Cell Dev Biol, 8, 2020
|
|
1Y51
 
 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) F29W mutant | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
1Y6I
 
 | | Synechocystis GUN4 | | Descriptor: | Mg-chelatase cofactor GUN4 | | Authors: | Verdecia, M.A, Larkin, R.M, Ferrer, J.L, Riek, R, Chory, J, Noel, J.P. | | Deposit date: | 2004-12-06 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Mg-chelatase cofactor GUN4 reveals a novel hand-shaped fold for porphyrin binding
Plos Biol., 3, 2005
|
|
1YNV
 
 | | Asp79 makes a large, unfavorable contribution to the stability of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Trevino, S.R, Gokulan, K, Newsom, S, Thurlkill, R.L, Shaw, K.L, Mitkevich, V.A, Makarov, A.A, Sacchettini, J.C, Scholtz, J.M, Pace, C.N. | | Deposit date: | 2005-01-25 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Asp79 Makes a Large, Unfavorable Contribution to the Stability of RNase Sa.
J.Mol.Biol., 354, 2005
|
|
1YRV
 
 | | Novel Ubiquitin-Conjugating Enzyme | | Descriptor: | ubiquitin-conjugating ligase MGC351130 | | Authors: | Walker, J.R, Choe, J, Avvakumov, G.V, Newman, E.M, MacKenzie, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-02-04 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
9HSN
 
 | |
9HSG
 
 | |
9HSJ
 
 | |
9HSS
 
 | |
9HSB
 
 | |
9HSO
 
 | |
