1NP3
 
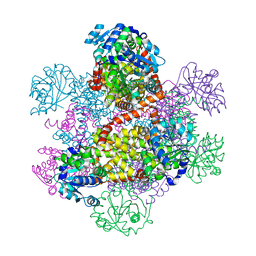 | | Crystal structure of class I acetohydroxy acid isomeroreductase from Pseudomonas aeruginosa | | Descriptor: | Ketol-acid reductoisomerase | | Authors: | Ahn, H.J, Eom, S.J, Yoon, H.-J, Lee, B.I, Cho, H, Suh, S.W. | | Deposit date: | 2003-01-17 | | Release date: | 2003-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Class I Acetohydroxy Acid Isomeroreductase from Pseudomonas aeruginosa
J.Mol.Biol., 328, 2003
|
|
2NA1
 
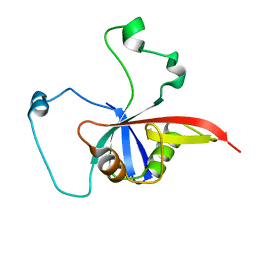 | | ULD complex | | Descriptor: | Polycomb complex protein BMI-1, Polyhomeotic-like 2 | | Authors: | Cierpicki, T, Gray, F, Cho, H. | | Deposit date: | 2015-12-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | BMI1 regulates PRC1 architecture and activity through homo- and hetero-oligomerization.
Nat Commun, 7, 2016
|
|
4B18
 
 | |
2XHS
 
 | |
4D3C
 
 | |
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
2X29
 
 | | Crystal structure of human4-1BB ligand ectodomain | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 9 | | Authors: | Won, E.Y, Cho, H.S. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of the Trimer of Human 4-1Bb Ligand is Unique Among Members of the Tumor Necrosis Factor Superfamily.
J.Biol.Chem., 285, 2010
|
|
5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
6DN2
 
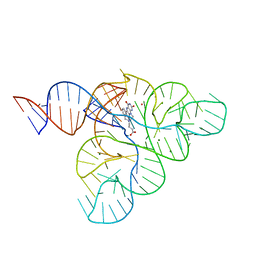 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1354 SPLIT RNA | | Descriptor: | 4-{benzyl[2-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)ethyl]amino}butanoic acid, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
6DN3
 
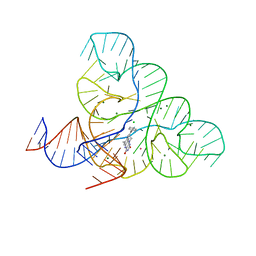 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1555 SPLIT RNA | | Descriptor: | 7,8-dimethyl-2,4-dioxo-10-(3-phenylpropyl)-1,2,3,4-tetrahydrobenzo[g]pteridin-10-ium, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
6DN1
 
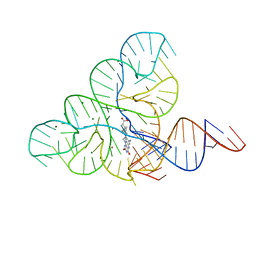 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1151 SPLIT RNA | | Descriptor: | 10-(6-carboxyhexyl)-8-(cyclopentylamino)-2,4-dihydroxy-7-methylbenzo[g]pteridin-10-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
1X1P
 
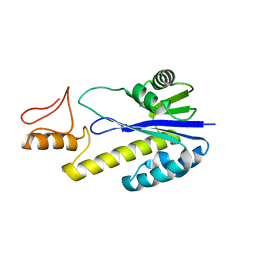 | | Crystal structure of Tk-RNase HII(1-197)-A(28-42) | | Descriptor: | Ribonuclease HII | | Authors: | Takano, K, Endo, S, Mukaiyama, A, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2005-04-11 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of amyloid beta fragments in aqueous environments
Febs J., 273, 2006
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
6XJI
 
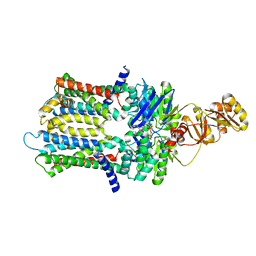 | | PmtCD ABC exporter at C1 symmetry | | Descriptor: | ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zeytuni, N, Strynadka, N.J.C, Hu, J, Worrall, L.J, Chou, H, Yu, Z. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
8T7C
 
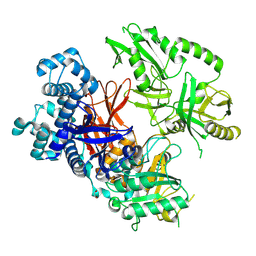 | | Crystal structure of human phospholipase C gamma 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2, CALCIUM ION | | Authors: | Chen, Y, Choi, H, Zhuang, N, Hu, L, Qian, D, Wang, J. | | Deposit date: | 2023-06-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal and cryo-EM structures of PLCg2 reveal dynamic inter-domain recognitions in autoinhibition
To Be Published
|
|
6XJH
 
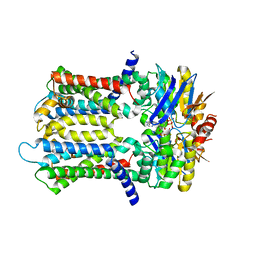 | | PmtCD ABC exporter without the basket domain at C2 symmetry | | Descriptor: | ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zeytuni, N, Strynadka, N.J.C, Hu, J, Worrall, L.J, Chou, H, Yu, Z. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
1LFW
 
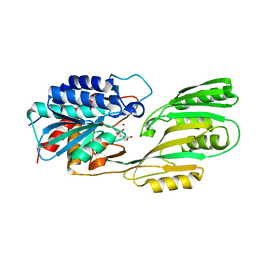 | | Crystal structure of pepV | | Descriptor: | 3-[(1-AMINO-2-CARBOXY-ETHYL)-HYDROXY-PHOSPHINOYL]-2-METHYL-PROPIONIC ACID, ZINC ION, pepV | | Authors: | Jozic, D, Bourenkow, G, Bartunik, H, Scholze, H, Dive, V, Henrich, B, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2002-04-12 | | Release date: | 2002-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Dinuclear Zinc Aminopeptidase PepV from Lactobacillus delbrueckii Unravels Its Preference for Dipeptides
Structure, 10, 2002
|
|
1KWF
 
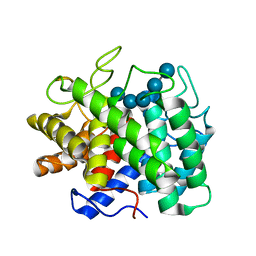 | | Atomic Resolution Structure of an Inverting Glycosidase in Complex with Substrate | | Descriptor: | Endoglucanase A, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Guerin, D.M.A, Lascombe, M.-B, Costabel, M, Souchon, H, Lamzin, V, Beguin, P, Alzari, P.M. | | Deposit date: | 2002-01-29 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Atomic (0.94 A) resolution structure of an inverting glycosidase in complex with substrate.
J.Mol.Biol., 316, 2002
|
|
2FH1
 
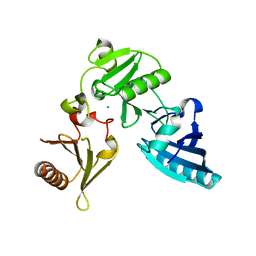 | | C-terminal half of gelsolin soaked in low calcium at pH 4.5 | | Descriptor: | CALCIUM ION, Gelsolin | | Authors: | Chumnarnsilpa, S, Loonchanta, A, Xue, B, Choe, H, Urosev, D, Wang, H, Burtnick, L.D, Robinson, R.C. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Calcium ion exchange in crystalline gelsolin
J.Mol.Biol., 357, 2006
|
|
3O3F
 
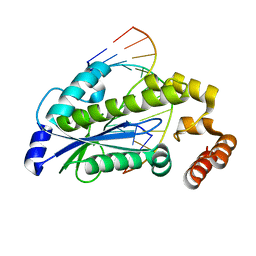 | | T. maritima RNase H2 D107N in complex with nucleic acid substrate and magnesium ions | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), DNA/RNA (5'-D(*GP*AP*CP*AP*C)-R(P*C)-D(P*TP*GP*AP*TP*TP*C)-3'), MAGNESIUM ION, ... | | Authors: | Rychlik, M.P, Chon, H, Cerritelli, S.M, Klimek, P, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-07-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.
Mol.Cell, 40, 2010
|
|
3O3H
 
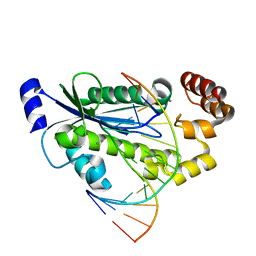 | | T. maritima RNase H2 D107N in complex with nucleic acid substrate and manganese ions | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), DNA/RNA (5'-D(*GP*AP*CP*AP*C)-R(P*C)-D(P*TP*GP*AP*TP*TP*C)-3'), MANGANESE (II) ION, ... | | Authors: | Rychlik, M.P, Chon, H, Cerritelli, S.M, Klimek, P, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-07-24 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of RNase H2 in Complex with Nucleic Acid Reveal the Mechanism of RNA-DNA Junction Recognition and Cleavage.
Mol.Cell, 40, 2010
|
|
2FH4
 
 | | C-terminal half of gelsolin soaked in EGTA at pH 8 | | Descriptor: | Gelsolin | | Authors: | Chumnarnsilpa, S, Loonchanta, A, Xue, B, Choe, H, Urosev, D, Wang, H, Burtnick, L.D, Robinson, R.C. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Calcium ion exchange in crystalline gelsolin
J.Mol.Biol., 357, 2006
|
|
2DFH
 
 | | Crystal structure of Tk-RNase HII(1-212)-C | | Descriptor: | Ribonuclease HII | | Authors: | Katagiri, Y, Takano, K, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2006-03-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Conformational contagion in a protein: Structural properties of a chameleon sequence
Proteins, 68, 2007
|
|
2DF5
 
 | | Crystal Structure of Pf-PCP(1-204)-C | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Katagiri, Y, Takano, K, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2006-02-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational contagion in a protein: structural properties of a chameleon sequence
Proteins, 68, 2007
|
|
2DFI
 
 | | Crystal structure of Pf-MAP(1-292)-C | | Descriptor: | COBALT (II) ION, Methionine aminopeptidase | | Authors: | Katagiri, Y, Takano, K, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2006-03-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational contagion in a protein: Structural properties of a chameleon sequence
Proteins, 68, 2007
|
|
