5YB9
 
 | | Crystal structure of a dimeric cyclophilin A from T.vaginalis | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Cho, C.C, Lin, M.H, Chou, C.C, Martin, T, Chen, C, Hsu, C.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | Structural basis of interaction between dimeric cyclophilin 1 and Myb1 transcription factor in Trichomonas vaginalis
Sci Rep, 8, 2018
|
|
5YBA
 
 | | Dimeric Cyclophilin from T.vaginalis in complex with Myb1 peptide | | Descriptor: | Myb1 peptide, Peptidyl-prolyl cis-trans isomerase | | Authors: | Cho, C.C, Lin, M.H, Martin, T, Chou, C.C, Chen, C, Hsu, C.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Structural basis of interaction between dimeric cyclophilin 1 and Myb1 transcription factor in Trichomonas vaginalis
Sci Rep, 8, 2018
|
|
8H3H
 
 | | Human ATAD2 Walker B mutant, ATP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family AAA domain-containing protein 2 | | Authors: | Cho, C, Song, J. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication.
Commun Biol, 6, 2023
|
|
5ZDC
 
 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P32) | | Descriptor: | PHOSPHATE ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE, poly ADP-ribose glycohydrolase | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDB
 
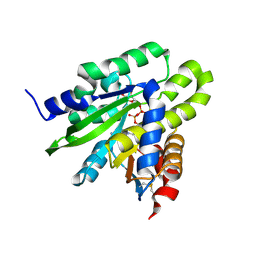 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P21) | | Descriptor: | Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDE
 
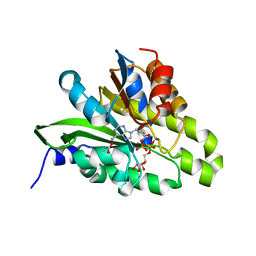 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P3221) | | Descriptor: | Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDA
 
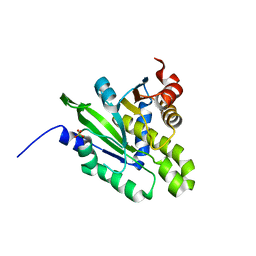 | |
5ZDD
 
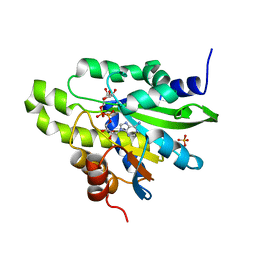 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P212121) | | Descriptor: | PHOSPHATE ION, Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.725 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDF
 
 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) T267K mutant from Deinococcus radiodurans in complex with ADP-ribose | | Descriptor: | Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDG
 
 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) T267R mutant from Deinococcus radiodurans in complex with ADP-ribose | | Descriptor: | Poly APD-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5DUS
 
 | | Crystal structure of MERS-CoV macro domain in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, ORF1a, ... | | Authors: | Cho, C.-C, Lin, M.-H, Chuang, C.-Y, Hsu, C.-H. | | Deposit date: | 2015-09-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Macro Domain from Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Is an Efficient ADP-ribose Binding Module: CRYSTAL STRUCTURE AND BIOCHEMICAL STUDIES
J.Biol.Chem., 291, 2016
|
|
6JQ0
 
 | | CryoEM structure of Abo1 Walker B (E372Q) mutant hexamer - ATP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Uncharacterized AAA domain-containing protein C31G5.19, ... | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-28 | | Release date: | 2019-12-25 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
2MRD
 
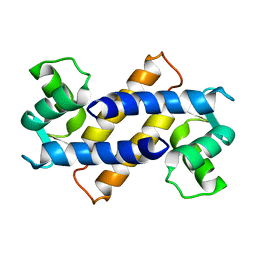 | |
6JPQ
 
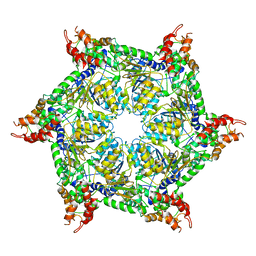 | | CryoEM structure of Abo1 hexamer - ADP complex | | Descriptor: | Uncharacterized AAA domain-containing protein C31G5.19 | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.44 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
6JPU
 
 | | CryoEM structure of Abo1 hexamer - apo complex | | Descriptor: | Uncharacterized AAA domain-containing protein C31G5.19 | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-28 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
8JUW
 
 | | Human ATAD2 Walker B mutant-H3/H4K5Q complex, ATP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family AAA domain-containing protein 2 | | Authors: | Cho, C, Song, J. | | Deposit date: | 2023-06-27 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication.
Commun Biol, 6, 2023
|
|
8JUZ
 
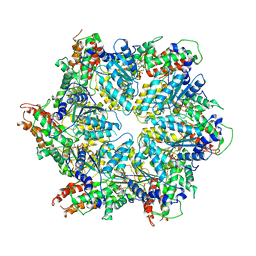 | | Human ATAD2 Walker B mutant-H3/H4K5Q complex, ATP state (Class III) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family AAA domain-containing protein 2 | | Authors: | Cho, C, Song, J. | | Deposit date: | 2023-06-27 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication.
Commun Biol, 6, 2023
|
|
8JUY
 
 | | Human ATAD2 Walker B mutant-H3/H4K5Q complex, ATP state (Class II) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family AAA domain-containing protein 2 | | Authors: | Cho, C, Song, J. | | Deposit date: | 2023-06-27 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication.
Commun Biol, 6, 2023
|
|
7X9D
 
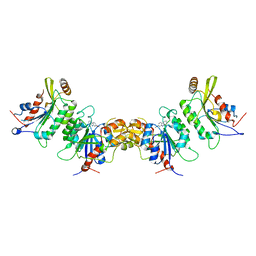 | | DNMT3B in complex with harmine | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B | | Authors: | Cho, C.-C, Yuan, H.S. | | Deposit date: | 2022-03-15 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Mechanistic Insights into Harmine-Mediated Inhibition of Human DNA Methyltransferases and Prostate Cancer Cell Growth.
Acs Chem.Biol., 18, 2023
|
|
8C8J
 
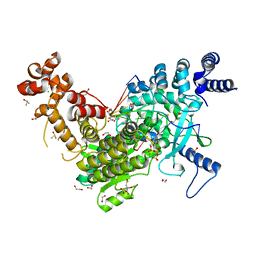 | | Long Interspersed Nuclear Element 1 (LINE-1) reverse transcriptase ternary complex with hybrid duplex and dTTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, CHLORIDE ION, ... | | Authors: | Nichols, C.E, Walpole, T.B, Baldwin, E. | | Deposit date: | 2023-01-20 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures, functions and adaptations of the human LINE-1 ORF2 protein.
Nature, 626, 2024
|
|
3QMZ
 
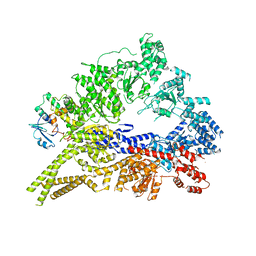 | | Crystal structure of the cytoplasmic dynein heavy chain motor domain | | Descriptor: | Cytoplasmic dynein heavy chain, Glutathione-S-transferase | | Authors: | Cho, C, Carter, A.P, Jin, L, Vale, R.D. | | Deposit date: | 2011-02-07 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal structure of the dynein motor domain.
Science, 331, 2011
|
|
2RCN
 
 | |
3LCW
 
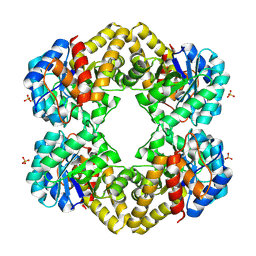 | | L-KDO aldolase complexed with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-12 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
2GEM
 
 | |
2GEL
 
 | |
