4V94
 
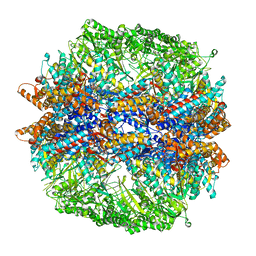 | | Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Leitner, A, Joachimiak, L.A, Bracher, A, Walzthoeni, T, Chen, B, Monkemeyer, L, Pechmann, S, Holmes, S, Cong, Y, Ma, B, Ludtke, S, Chiu, W, Hartl, F.U, Aebersold, R, Frydman, J. | | Deposit date: | 2012-01-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT.
Structure, 20, 2012
|
|
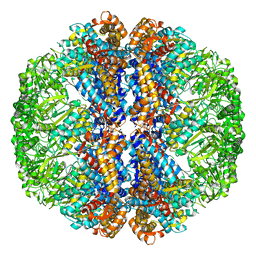
3J3X
 
 | |
1R0M
 
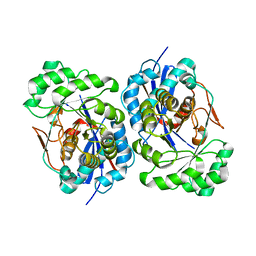 | | Structure of Deinococcus radiodurans N-acylamino acid racemase at 1.3 : insights into a flexible binding pocket and evolution of enzymatic activity | | Descriptor: | N-acylamino acid racemase | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
3ZQ0
 
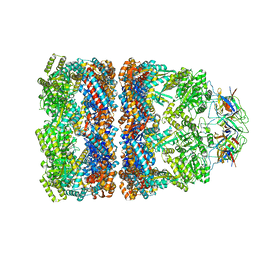 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | Descriptor: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
1QVR
 
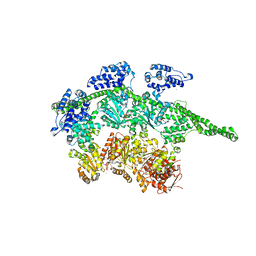 | | Crystal Structure Analysis of ClpB | | Descriptor: | ClpB protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLATINUM (II) ION | | Authors: | Lee, S, Sowa, M.E, Watanabe, Y, Sigler, P.B, Chiu, W, Yoshida, M, Tsai, F.T.F. | | Deposit date: | 2003-08-28 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of ClpB: A Molecular Chaperone that Rescues Proteins from an Aggregated State
Cell(Cambridge,Mass.), 115, 2003
|
|
7LIH
 
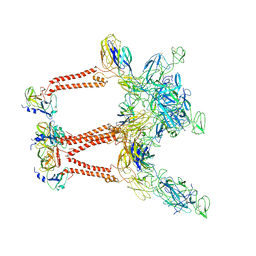 | | CryoEM structure of Mayaro virus icosahedral subunit | | Descriptor: | Capsid protein, E1 protein, E2 protein | | Authors: | Chmielewski, D, Kaelber, J.T, Jin, J, Weaver, S, Auguste, A.J, Chiu, W. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Near-atomic resolution Cryo-EM structure of Mayaro virus identifies key structural determinants of alphavirus particle formation
To Be Published
|
|
4A0O
 
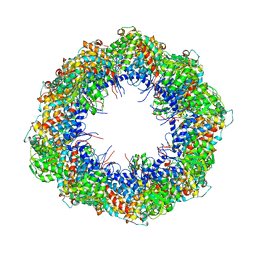 | | Symmetry-free cryo-EM map of TRiC in the nucleotide-free (apo) state | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
6M0R
 
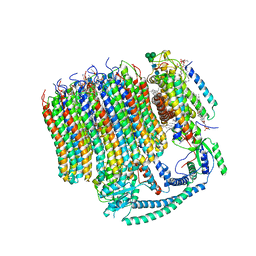 | | 2.7A Yeast Vo state3 | | Descriptor: | (6~{E},10~{E},14~{E},18~{E},22~{E},26~{E},30~{R})-2,6,10,14,18,22,26,30-octamethyldotriaconta-2,6,10,14,18,22,26-heptaene, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, PYROPHOSPHATE, ... | | Authors: | Roh, S.H, Shekhar, M, Pintilie, G, Chipot, C, Wilkens, S, Singharoy, A, Chiu, W. | | Deposit date: | 2020-02-22 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM and MD infer water-mediated proton transport and autoinhibition mechanisms of V o complex.
Sci Adv, 6, 2020
|
|
6UKT
 
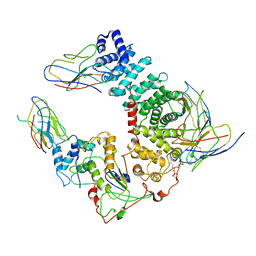 | | Cryo-EM structure of mammalian Ric-8A:Galpha(i):nanobody complex | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, NB8109, NB8117, ... | | Authors: | Mou, T.C, Zhang, K, Johnston, J.D, Chiu, W, Sprang, S.R. | | Deposit date: | 2019-10-05 | | Release date: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structure of the G protein chaperone and guanine nucleotide exchange factor Ric-8A bound to G alpha i1.
Nat Commun, 11, 2020
|
|
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | Descriptor: | RNA (387-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSM
 
 | | Misfolded Tetrahymena ribozyme conformation 3 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSL
 
 | | Misfolded Tetrahymena ribozyme conformation 2 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSK
 
 | | Misfolded Tetrahymena ribozyme conformation 1 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5NG5
 
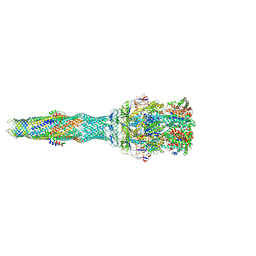 | | multi-drug efflux; membrane transport; RND superfamily; Drug resistance | | Descriptor: | 6-[2-(3,4-dimethoxyphenyl)ethylsulfanyl]-8-[4-(2-methoxyethyl)piperazin-1-yl]-3,3-dimethyl-1,4-dihydropyrano[3,4-c]pyridine-5-carbonitrile, Multidrug efflux pump accessory protein AcrZ, Multidrug efflux pump subunit AcrA, ... | | Authors: | Wang, Z, Fan, G, Hryc, C.F, Blaza, J.N, Serysheva, I.I, Schmid, M.F, Chiu, W, Luisi, B.F, Du, D. | | Deposit date: | 2017-03-16 | | Release date: | 2017-04-19 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | An allosteric transport mechanism for the AcrAB-TolC Multidrug Efflux Pump.
Elife, 6, 2017
|
|
7WU7
 
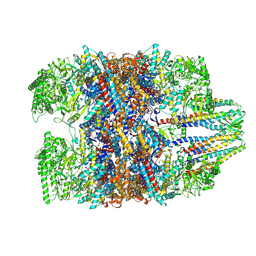 | | Prefoldin-tubulin-TRiC complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Prefoldin subunit 1, Prefoldin subunit 2, ... | | Authors: | Gestaut, D, Zhao, Y, Park, J, Ma, B, Leitner, A, Collier, M, Pintilie, G, Roh, S.-H, Chiu, W, Frydman, J. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
6M0S
 
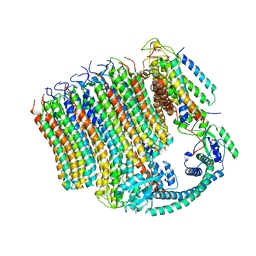 | | 3.6A Yeast Vo state3 prime | | Descriptor: | Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, vacuolar isoform, ... | | Authors: | Roh, S.H, Shekhar, M, Pintilie, G, Chipot, C, Wilkens, S, SIngharoy, A, Chiu, W. | | Deposit date: | 2020-02-22 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM and MD infer water-mediated proton transport and autoinhibition mechanisms of V o complex.
Sci Adv, 6, 2020
|
|
6C54
 
 | | Ebola nucleoprotein nucleocapsid-like assembly and the asymmetric unit | | Descriptor: | Nucleoprotein | | Authors: | Su, Z, Wu, C, Pintilie, G.D, Chiu, W, Amarasinghe, G.K, Leung, D.W. | | Deposit date: | 2018-01-13 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Electron Cryo-microscopy Structure of Ebola Virus Nucleoprotein Reveals a Mechanism for Nucleocapsid-like Assembly.
Cell, 172, 2018
|
|
8FR7
 
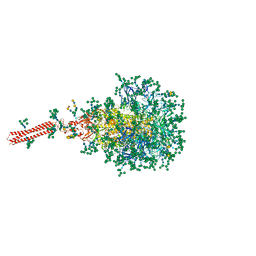 | | A hinge glycan regulates spike bending and impacts coronavirus infectivity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pintilie, G, Wilson, E, Chmielewski, D, Schmid, M.F, Jin, J, Chen, M, Singharoy, A, Chiu, W. | | Deposit date: | 2023-01-06 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural insights into the modulation of coronavirus spike tilting and infectivity by hinge glycans.
Nat Commun, 14, 2023
|
|
7RAK
 
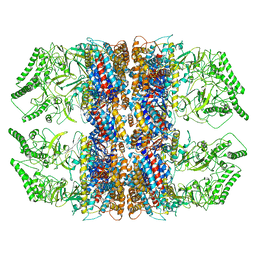 | | Methanococcus maripaludis chaperonin complex in open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-07-01 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7RRP
 
 | |
7TBH
 
 | | cryo-EM structure of MBP-KIX-apoferritin complex with peptide 7 | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed, LEU-SER-ARG-ARG-PRO-SEP-TYR-ARG-LYS-ILE-LEU-ASN-ASP-LEU-SER-SER-ASP-ALA-PRO | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
6WLM
 
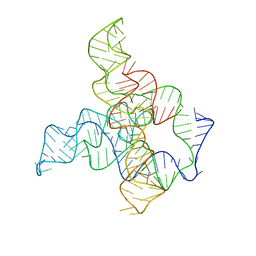 | | F. nucleatum glycine riboswitch with glycine models, 7.4 Angstrom resolution | | Descriptor: | RNA (171-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLR
 
 | | SAM-IV riboswitch with SAM models, 4.8 Angstrom resolution | | Descriptor: | RNA (119-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLS
 
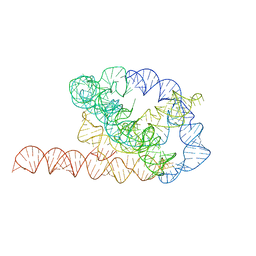 | | Tetrahymena ribozyme models, 6.8 Angstrom resolution | | Descriptor: | RNA (388-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
7TB3
 
 | | cryo-EM structure of MBP-KIX-apoferritin | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
