6TCI
 
 | |
5JIP
 
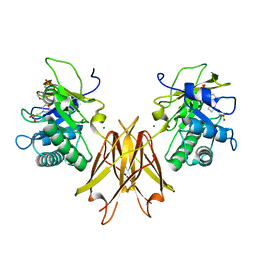 | | Crystal structure of the Clostridium perfringens spore cortex lytic enzyme SleM | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cortical-lytic enzyme, MAGNESIUM ION | | 著者 | Chirgadze, D.Y, Christie, G, Ustok, F.I, Al-Riyami, B, Stott, K. | | 登録日 | 2016-04-22 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of Clostridium perfringens SleM, a muramidase involved in cortical hydrolysis during spore germination.
Proteins, 84, 2016
|
|
1U41
 
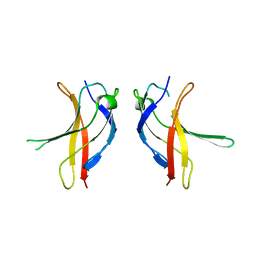 | | Crystal structure of YLGV mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-23 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U36
 
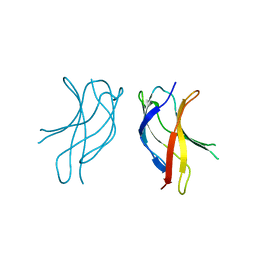 | | Crystal structure of WLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-21 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3Y
 
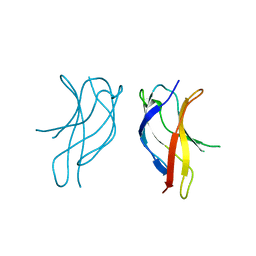 | | Crystal structure of ILAC mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-22 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U42
 
 | | Crystal structure of MLAM mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-23 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.699 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3Z
 
 | | Crystal structure of MLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-23 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3J
 
 | | Crystal structure of MLAV mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-22 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1NK1
 
 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | 分子名称: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | 著者 | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | 登録日 | 1998-08-20 | | 公開日 | 1999-01-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
3KX8
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK | | 分子名称: | fluoroacetyl-CoA thioesterase | | 著者 | Chirgadze, D.Y, Dias, M.V.B, Huang, F, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | 登録日 | 2009-12-02 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3G04
 
 | | Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HUMAN THYROID STIMULATING AUTOANTIBODY M22 HEAVY CHAIN, HUMAN THYROID STIMULATING AUTOANTIBODY M22 LIGHT CHAIN, ... | | 著者 | Sanders, J, Chirgadze, D.Y, Sanders, P, Baker, S, Sullivan, A, Bhardwaja, A, Bolton, J, Reeve, M, Nakatake, N, Evans, M, Richards, T, Powell, M, Miguel, R.N, Blundell, T.L, Furmaniak, J, Smith, B.R. | | 登録日 | 2009-01-27 | | 公開日 | 2009-08-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody
Thyroid, 17, 2007
|
|
6T1H
 
 | | OXA-51-like beta-lactamase OXA-66 | | 分子名称: | Beta-lactamase OXA-66, ZINC ION | | 著者 | Takebayashi, Y, Chirgadze, D, Henderson, S, Warburton, P.J, Evans, B.E. | | 登録日 | 2019-10-04 | | 公開日 | 2020-10-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the OXA-51-like beta-lactamase OXA-66
To Be Published
|
|
6ZHA
 
 | | Cryo-EM structure of DNA-PK monomer | | 分子名称: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.91 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH2
 
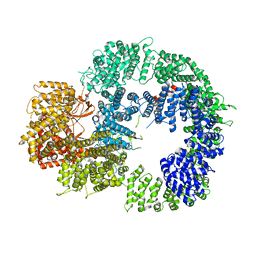 | | Cryo-EM structure of DNA-PKcs (State 1) | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-20 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH8
 
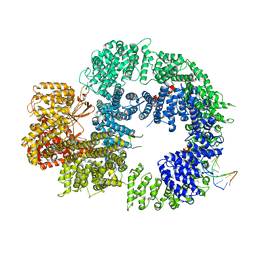 | | Cryo-EM structure of DNA-PKcs:DNA | | 分子名称: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*T)-3'), DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH6
 
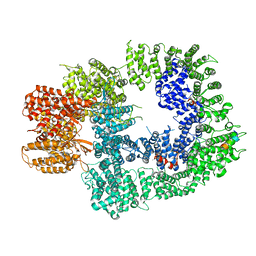 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH4
 
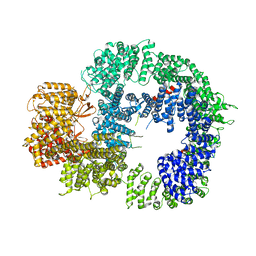 | | Cryo-EM structure of DNA-PKcs (State 3) | | 分子名称: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-20 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5CSQ
 
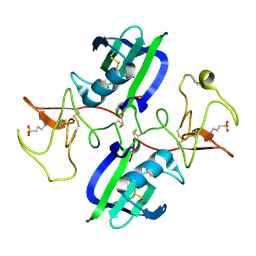 | | The structure of the NK1 fragment of HGF/SF complexed with MOPS | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Hepatocyte growth factor | | 著者 | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | 登録日 | 2015-07-23 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CS9
 
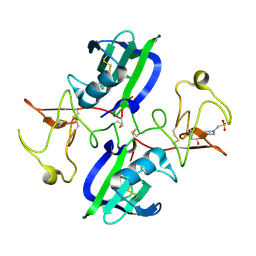 | | The structure of the NK1 fragment of HGF/SF complexed with MES | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hepatocyte growth factor | | 著者 | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | 登録日 | 2015-07-23 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CT1
 
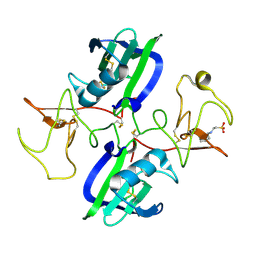 | | The structure of the NK1 fragment of HGF/SF complexed with CHES | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hepatocyte growth factor | | 著者 | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | 登録日 | 2015-07-23 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CS1
 
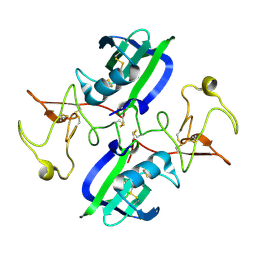 | | The structure of the NK1 fragment of HGF/SF | | 分子名称: | Hepatocyte growth factor | | 著者 | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | 登録日 | 2015-07-23 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CS5
 
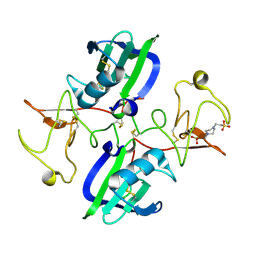 | | The structure of the NK1 fragment of HGF/SF complexed with PIPES | | 分子名称: | Hepatocyte growth factor, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID) | | 著者 | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | 登録日 | 2015-07-23 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CT2
 
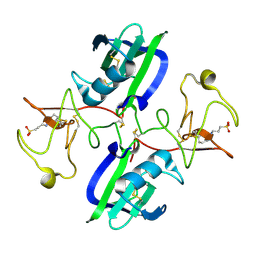 | | The structure of the NK1 fragment of HGF/SF complexed with CAPS | | 分子名称: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Hepatocyte growth factor | | 著者 | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | 登録日 | 2015-07-23 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
7NFE
 
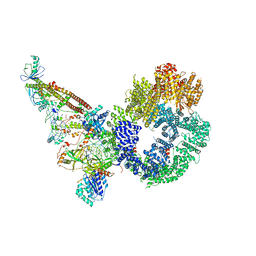 | | Cryo-EM structure of NHEJ super-complex (monomer) | | 分子名称: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*CP*TP*AP*TP*TP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(P*TP*AP*AP*TP*AP*AP*TP*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*AP*G)-3'), DNA ligase 4, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2021-02-06 | | 公開日 | 2021-08-18 | | 最終更新日 | 2021-09-22 | | 実験手法 | ELECTRON MICROSCOPY (4.29 Å) | | 主引用文献 | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
7NFC
 
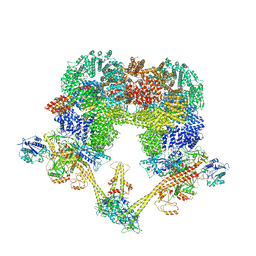 | | Cryo-EM structure of NHEJ super-complex (dimer) | | 分子名称: | DNA (27-MER), DNA (28-MER), DNA ligase 4, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2021-02-05 | | 公開日 | 2021-08-18 | | 最終更新日 | 2021-09-22 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
