1NHA
 
 | | Solution Structure of the Carboxyl-Terminal Domain of RAP74 and NMR Characterization of the FCP-Binding Sites of RAP74 and CTD of RAP74, the subunit of Human TFIIF | | Descriptor: | Transcription initiation factor IIF, alpha subunit | | Authors: | Nguyen, B.D, Chen, H.T, Kobor, M.S, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Carboxyl-Terminal Domain of RAP74 and NMR Characterization of the FCP1-Binding Sites of RAP74 and Human TFIIB.
Biochemistry, 42, 2003
|
|
2GS0
 
 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the activation domain of p53 | | Descriptor: | Cellular tumor antigen p53, RNA polymerase II transcription factor B subunit 1 | | Authors: | Di Lello, P, Jones, T.N, Nguyen, B.D, Legault, P, Omichinski, J.G. | | Deposit date: | 2006-04-25 | | Release date: | 2006-10-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53.
Mol.Cell, 22, 2006
|
|
1ETZ
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF AN ANTI-SWEETENER FAB, NC10.14, SHOWS THE EXTENT OF STRUCTURAL DIVERSITY IN ANTIGEN RECOGNITION BY IMMUNOGLOBULINS | | Descriptor: | FAB NC10.14 - HEAVY CHAIN, FAB NC10.14 - LIGHT CHAIN, N-(P-CYANOPHENYL)-N'-DIPHENYLMETHYL-GUANIDINE-ACETIC ACID | | Authors: | Guddat, L.W, Shan, L, Broomell, C, Ramsland, P.A, Fan, Z, Anchin, J.M, Linthicum, D.S, Edmundson, A.B. | | Deposit date: | 2000-04-13 | | Release date: | 2000-10-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The three-dimensional structure of a complex of a murine Fab (NC10. 14) with a potent sweetener (NC174): an illustration of structural diversity in antigen recognition by immunoglobulins.
J.Mol.Biol., 302, 2000
|
|
8UQT
 
 | | Crystal structure of the Tree Shrew p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53, SULFATE ION | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
8UQS
 
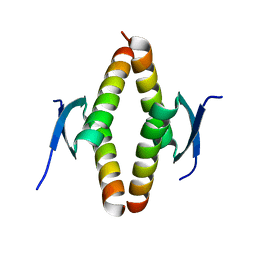 | | Crystal structure of the Opossum p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53 (Fragment) | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
8UQR
 
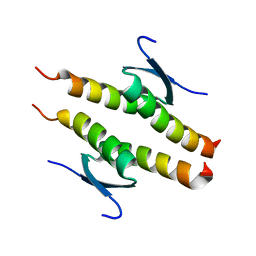 | | Crystal structure of the human p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53 | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
1ONV
 
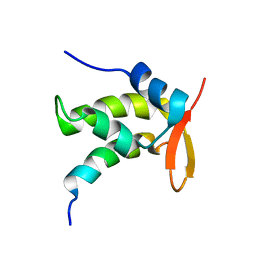 | | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNAP II CTD Phosphatase FCP1 | | Descriptor: | Transcription initiation factor IIF, alpha subunit, serine phosphatase FCP1a | | Authors: | Nguyen, B.D, Abbott, K.L, Potempa, K, Kobor, M.S, Archambault, J, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNA polymerase II carboxyl-terminal domain phosphatase FCP1
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1OLH
 
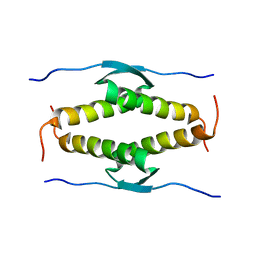 | |
1S6L
 
 | | Solution structure of MerB, the Organomercurial Lyase involved in the bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase | | Authors: | Di Lello, P, Benison, G.C, Valafar, H, Pitts, K.E, Summers, A.O, Legault, P, Omichinski, J.G. | | Deposit date: | 2004-01-25 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies reveal a novel protein fold for MerB, the organomercurial lyase involved in the bacterial mercury resistance system.
Biochemistry, 43, 2004
|
|
1CQW
 
 | | NAI COCRYSTALLISED WITH HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | HALOALKANE DEHALOGENASE; 1-CHLOROHEXANE HALIDOHYDROLASE, IODIDE ION | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1999-08-11 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
4H9K
 
 | | Crystal structure of cleavage site mutant of Npro of classical swine fever virus. | | Descriptor: | Hog cholera virus, SULFATE ION, ZINC ION | | Authors: | Gottipati, K, Ruggli, N, Gerber, M, Tratschin, J.-D, Benning, M, Bellamy, H, Choi, K.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The Structure of Classical Swine Fever Virus N(pro): A Novel Cysteine Autoprotease and Zinc-Binding Protein Involved in Subversion of Type I Interferon Induction.
Plos Pathog., 9, 2013
|
|
1DL6
 
 | | SOLUTION STRUCTURE OF HUMAN TFIIB N-TERMINAL DOMAIN | | Descriptor: | TRANSCRIPTION FACTOR II B (TFIIB), ZINC ION | | Authors: | Chen, H.-T, Legault, P, Glushka, J, Omichinski, J.G, Scott, R.A. | | Deposit date: | 1999-12-08 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a (Cys3His) zinc ribbon, a ubiquitous motif in archaeal and eucaryal transcription.
Protein Sci., 9, 2000
|
|
2GAT
 
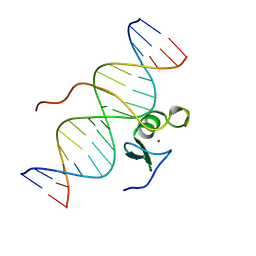 | | SOLUTION STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
2K2U
 
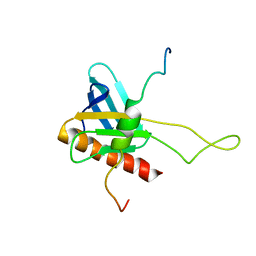 | | NMR Structure of the complex between Tfb1 subunit of TFIIH and the activation domain of VP16 | | Descriptor: | Alpha trans-inducing protein, RNA polymerase II transcription factor B subunit 1 | | Authors: | Langlois, C, Mas, C, Di Lello, P, Miller Jenkins, P.M, Legault, J, Omichinski, J.G. | | Deposit date: | 2008-04-11 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Complex between the Tfb1 Subunit of TFIIH and the Activation Domain of VP16: Structural Similarities between VP16 and p53.
J.Am.Chem.Soc., 130, 2008
|
|
1GAT
 
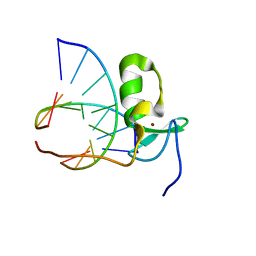 | | SOLUTION STRUCTURE OF THE SPECIFIC DNA COMPLEX OF THE ZINC CONTAINING DNA BINDING DOMAIN OF THE ERYTHROID TRANSCRIPTION FACTOR GATA-1 BY MULTIDIMENSIONAL NMR | | Descriptor: | DNA (5'-D(P*AP*GP*AP*TP*AP*AP*AP*C)3'), DNA (5'-D(P*GP*TP*TP*TP*AP*TP*CP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1993-06-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1.
Science, 261, 1993
|
|
1GAU
 
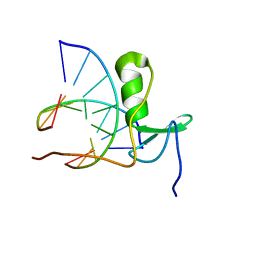 | |
4H9J
 
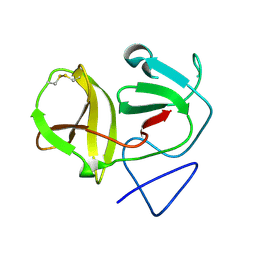 | | Crystal structure of N-terminal protease (Npro) of classical swine fever virus. | | Descriptor: | Hog cholera virus | | Authors: | Gottipati, K, Ruggli, N, Gerber, M, Tratschin, J.-D, Benning, M, Bellamy, H, Choi, K.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-10-30 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of Classical Swine Fever Virus N(pro): A Novel Cysteine Autoprotease and Zinc-Binding Protein Involved in Subversion of Type I Interferon Induction.
Plos Pathog., 9, 2013
|
|
1TNS
 
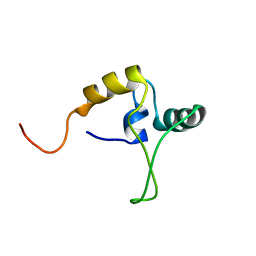 | |
1TNT
 
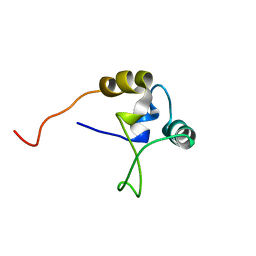 | |
2KDT
 
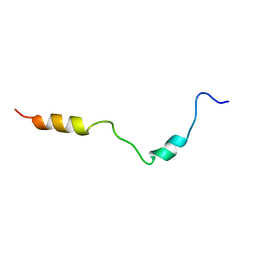 | | PC1/3 DCSG sorting domain structure in DPC | | Descriptor: | Neuroendocrine convertase 1 | | Authors: | Dikeakos, J.D, Di Lello, P, Lacombe, M.J, Ghirlando, R, Legault, P, Reudelhuber, T.L, Omichinski, J.G. | | Deposit date: | 2009-01-19 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Functional and structural characterization of a dense core secretory granule sorting domain from the PC1/3 protease.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1NCP
 
 | |
2JTX
 
 | |
2KE3
 
 | | PC1/3 DCSG sorting domain in CHAPS | | Descriptor: | Neuroendocrine convertase 1 | | Authors: | Dikeakos, J.D, Di Lello, P, Lacombe, M.J, Ghirlando, R, Legault, P, Reudelhuber, T.L, Omichinski, J.G. | | Deposit date: | 2009-01-22 | | Release date: | 2009-04-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Functional and structural characterization of a dense core secretory granule sorting domain from the PC1/3 protease
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2M14
 
 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and Rad4 | | Descriptor: | DNA repair protein RAD4, RNA polymerase II transcription factor B subunit 1 | | Authors: | Lafrance-Vanasse, J, Arseneault, G, Cappadocia, L, Legault, P, Omichinski, J.G. | | Deposit date: | 2012-11-16 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional evidence that Rad4 competes with Rad2 for binding to the Tfb1 subunit of TFIIH in NER.
Nucleic Acids Res., 41, 2013
|
|
2MBH
 
 | | NMR structure of EKLF(22-40)/Ubiquitin Complex | | Descriptor: | Krueppel-like factor 1, Ubiquitin | | Authors: | Raiola, L, Omichinski, J.G. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a Noncovalent Complex between Ubiquitin and the Transactivation Domain of the Erythroid-Specific Factor EKLF.
Structure, 21, 2013
|
|
