5SMG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2092370954 | | Descriptor: | 3-amino-N-ethyl-N-methylbenzamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1430613393 | | Descriptor: | 3-fluoro-N-(3-hydroxy-4-methylphenyl)benzamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SL8
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434762 | | Descriptor: | N,N-dimethylpyridin-4-amine, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SMH
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434938 | | Descriptor: | 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SM9
 
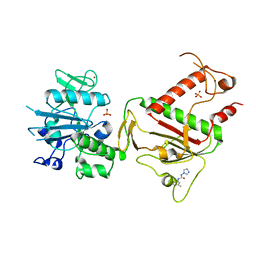 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2234920345 | | Descriptor: | N-(2-methoxy-5-methylphenyl)-N'-4H-1,2,4-triazol-4-ylurea, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2QH3
 
 | |
3MAY
 
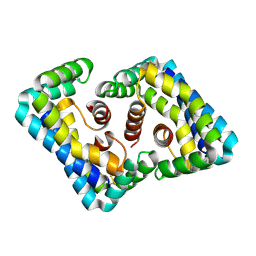 | |
4Y0L
 
 | | Mycobacterial membrane protein MmpL11D2 | | Descriptor: | IODIDE ION, Putative membrane protein mmpL11, SULFATE ION | | Authors: | Torres, R, Chim, N, Goulding, C.W. | | Deposit date: | 2015-02-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure and Interactions of Periplasmic Domains of Crucial MmpL Membrane Proteins from Mycobacterium tuberculosis.
Chem.Biol., 22, 2015
|
|
8T3X
 
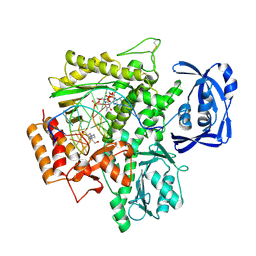 | | TNA polymerase, closed ternary | | Descriptor: | 10-92, TNA polymerase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Maola, V, Chaput, J, Chim, N. | | Deposit date: | 2023-06-07 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Directed evolution of a highly efficient TNA polymerase achieved by homologous recombination
Nat Catal, 2024
|
|
7TQW
 
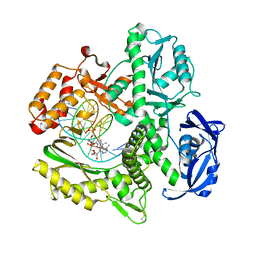 | | Kod RSGA incorporating PMT, n+2 | | Descriptor: | DNA polymerase, Primer, Template | | Authors: | Hajjar, M, Chim, N, Chaput, J.C. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystallographic analysis of engineered polymerases synthesizing phosphonomethylthreosyl nucleic acid.
Nucleic Acids Res., 50, 2022
|
|
6MU4
 
 | | Bst DNA polymerase I FANA/DNA binary complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA polymerase I, ... | | Authors: | Jackson, L.N, Chim, N, Chaput, J.C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of a natural DNA polymerase that functions as an XNA reverse transcriptase.
Nucleic Acids Res., 47, 2019
|
|
6MU5
 
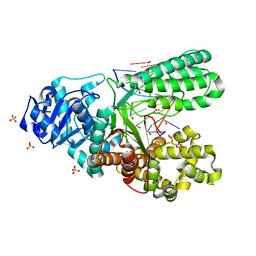 | | Bst DNA polymerase I TNA/DNA binary complex | | Descriptor: | DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA polymerase I, SULFATE ION, ... | | Authors: | Jackson, L.N, Chim, N, Chaput, J.C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | Crystal structures of a natural DNA polymerase that functions as an XNA reverse transcriptase.
Nucleic Acids Res., 47, 2019
|
|
4IVH
 
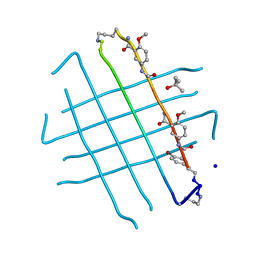 | | Crystal structure of QKLVFFAED nonapeptide segment from amyloid beta incorporated into a macrocyclic beta-sheet template | | Descriptor: | SODIUM ION, TERTIARY-BUTYL ALCOHOL, cyclo[Gln-Lys-Leu-Val-Phe-Phe-Ala-Glu-Asp-(delta-linked-Orn)-Hao-Lys-Hao-(p-bromoPhe)-Thr-(delta-linked-Orn)] | | Authors: | Pham, J.D, Chim, N, Goulding, C.W, Nowick, J.S. | | Deposit date: | 2013-01-22 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of beta-Amyloid Peptide Oligomers
J.Am.Chem.Soc., 2013
|
|
2LBX
 
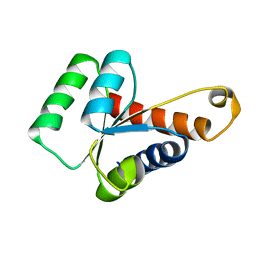 | | Solution structure of the S. cerevisiae H/ACA RNP protein Nhp2p | | Descriptor: | H/ACA ribonucleoprotein complex subunit 2 | | Authors: | Koo, B, Park, C, Fernandez, C.F, Chim, N, Ding, Y, Chanfreau, G, Feigon, J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of H/ACA RNP Protein Nhp2p Reveals Cis/Trans Isomerization of a Conserved Proline at the RNA and Nop10 Binding Interface.
J.Mol.Biol., 411, 2011
|
|
2LBW
 
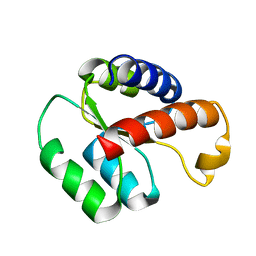 | | Solution structure of the S. cerevisiae H/ACA RNP protein Nhp2p-S82W mutant | | Descriptor: | H/ACA ribonucleoprotein complex subunit 2 | | Authors: | Koo, B, Park, C, Fernandez, C.F, Chim, N, Ding, Y, Chanfreau, G, Feigon, J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of H/ACA RNP Protein Nhp2p Reveals Cis/Trans Isomerization of a Conserved Proline at the RNA and Nop10 Binding Interface.
J.Mol.Biol., 411, 2011
|
|
2QH4
 
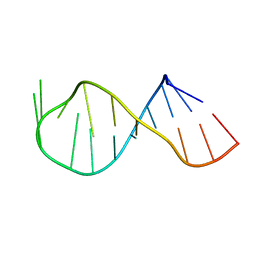 | |
2QH2
 
 | |
7RSU
 
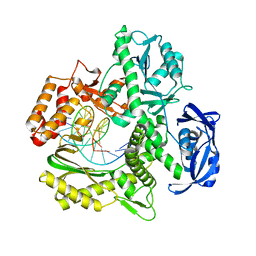 | | TNA polymerase, n+2 product | | Descriptor: | DNA polymerase, Primer, Template | | Authors: | Maola, V, Chaput, J, Chim, N. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis and Polymerase Recognition of Threose Nucleic Acid Triphosphates Equipped with Diverse Chemical Functionalities.
J.Am.Chem.Soc., 143, 2021
|
|
7RSS
 
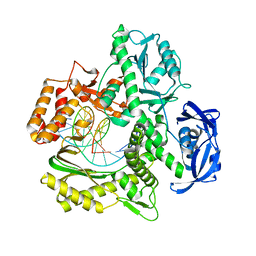 | | Kod-RI incorporating DNA, n+2 | | Descriptor: | DNA polymerase, Primer, Template | | Authors: | Hajjar, M, Chim, N, Chaput, J.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystallographic analysis of engineered polymerases synthesizing phosphonomethylthreosyl nucleic acid.
Nucleic Acids Res., 50, 2022
|
|
7RSR
 
 | | Kod-RI incorporating PMT, n+2 | | Descriptor: | DNA polymerase, Primer, Template | | Authors: | Hajjar, M, Chim, N, Chaput, J.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic analysis of engineered polymerases synthesizing phosphonomethylthreosyl nucleic acid.
Nucleic Acids Res., 50, 2022
|
|
7MCC
 
 | | Crystal structure of an AI-designed TIM-barrel F2C | | Descriptor: | AI-designed TIM-barrel F2C, SULFATE ION | | Authors: | Mathews, I.I, Anand-Achim, N, Perez, C.P, Huang, P.S. | | Deposit date: | 2021-04-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Protein sequence design with a learned potential.
Nat Commun, 13, 2022
|
|
7MCD
 
 | |
7SMJ
 
 | |
7QGI
 
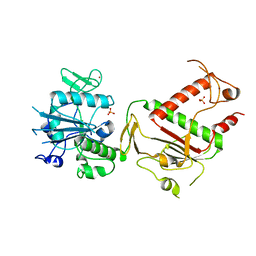 | | Crystal structure of SARS-CoV-2 NSP14 in the absence of NSP10 | | Descriptor: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-08 | | Release date: | 2022-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
7QIF
 
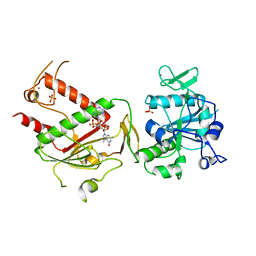 | | Crystal structure of SARS-CoV-2 NSP14 in complex with 7MeGpppG. | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
