3GYX
 
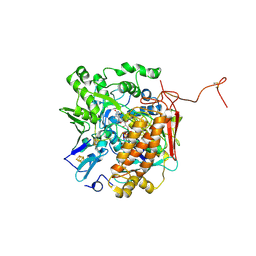 | | Crystal structure of adenylylsulfate reductase from Desulfovibrio gigas | | Descriptor: | Adenylylsulfate Reductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER | | Authors: | Chiang, Y.-L, Hsieh, Y.-C, Liu, E.-H, Liu, M.-Y, Chen, C.-J. | | Deposit date: | 2009-04-06 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Adenylylsulfate reductase from Desulfovibrio gigas suggests a potential self-regulation mechanism involving the C terminus of the beta-subunit
J.Bacteriol., 191, 2009
|
|
5FNY
 
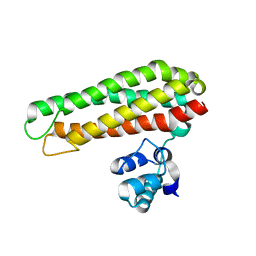 | | Low solvent content crystal form of Zn containing Iron sulfur cluster repair protein YtfE | | Descriptor: | FE (III) ION, IRON-SULFUR CLUSTER REPAIR PROTEIN YTFE, NONAETHYLENE GLYCOL, ... | | Authors: | Lo, F.-C, Hsieh, C.-C, Maestre-Reyna, M, Chen, C.-Y, Ko, T.-P, Horng, Y.-C, Lai, Y.-C, Chiang, Y.-W, Chou, C.-M, Chiang, C.-H, Huang, W.-N, Liaw, W.-F. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of the Repair of Iron Centers Protein Ytfe and its Interaction with No
Chemistry, 22, 2016
|
|
5FNP
 
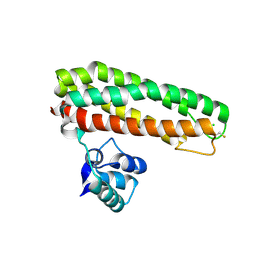 | | High resolution Zn containing Iron sulfur cluster repair protein YtfE | | Descriptor: | CHLORIDE ION, IRON-SULFUR CLUSTER REPAIR PROTEIN YTFE, OXYGEN ATOM, ... | | Authors: | Lo, F.-C, Hsieh, C.-C, Maestre-Reyna, M, Chen, C.-Y, Ko, T.-P, Horng, Y.-C, Lai, Y.-C, Chiang, Y.-W, Chou, C.-M, Chiang, C.-H, Huang, W.-N, Liaw, W.-F. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Repair of Iron Centers Protein Ytfe and its Interaction with No
Chemistry, 22, 2016
|
|
5FNN
 
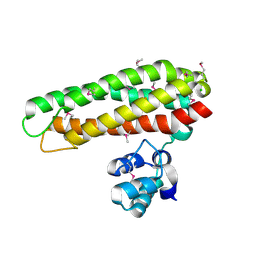 | | Iron and Selenomethionine containing Iron sulfur cluster repair protein YtfE | | Descriptor: | CHLORIDE ION, FE (II) ION, FE (III) ION, ... | | Authors: | Lo, F.-C, Hsieh, C.-C, Maestre-Reyna, M, Chen, C.-Y, Ko, T.-P, Horng, Y.-C, Lai, Y.-C, Chiang, Y.-W, Chou, C.-M, Chiang, C.-H, Huang, W.-N, Liaw, W.-F. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the Repair of Iron Centers Protein Ytfe and its Interaction with No
Chemistry, 22, 2016
|
|
6EEQ
 
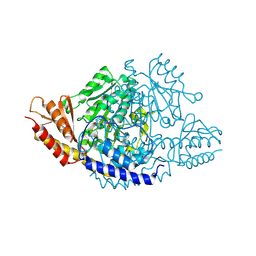 | | Crystal structure of Rhodiola rosea 4-hydroxyphenylacetaldehyde synthase | | Descriptor: | 4-hydroxyphenylacetaldehyde synthase | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.600086 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEI
 
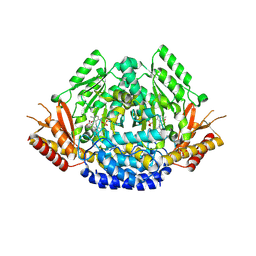 | | Crystal structure of Arabidopsis thaliana phenylacetaldehyde synthase in complex with L-phenylalanine | | Descriptor: | PHENYLALANINE, SULFATE ION, Tyrosine decarboxylase 1 | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-14 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (1.99001348 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEW
 
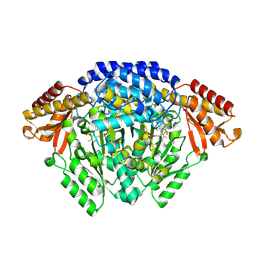 | | Crystal structure of Catharanthus roseus tryptophan decarboxylase in complex with L-tryptophan | | Descriptor: | Aromatic-L-amino-acid decarboxylase, CALCIUM ION, TRYPTOPHAN | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.05002069 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2KL7
 
 | |
3OR1
 
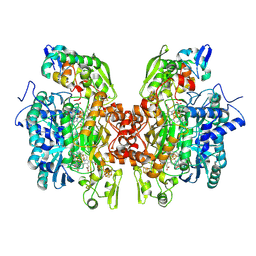 | | Crystal structure of dissimilatory sulfite reductase I (DsrI) | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Hsieh, Y.C, Liu, M.Y, Wang, V.C.C, Chiang, Y.L, Liu, E.H, Wu, W.G, Chan, S.I, Chen, C.J. | | Deposit date: | 2010-09-06 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure insights into the enzyme catalysis from comparison of three forms of dissimilatory sulfite reductase from Desulfovibrio gigas
To be Published
|
|
1Y7H
 
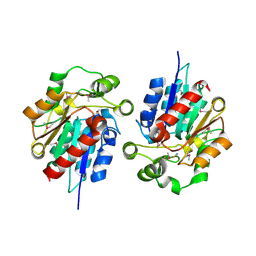 | | Structural and biochemical studies identify tobacco SABP2 as a methylsalicylate esterase and further implicate it in plant innate immunity, Northeast Structural Genomics Target AR2241 | | Descriptor: | THIOCYANATE ION, salicylic acid-binding protein 2 | | Authors: | Forouhar, F, Yang, Y, Kumar, D, Chen, Y, Fridman, E, Park, S.W, Chiang, Y, Acton, T.B, Montelione, G.T, Pichersky, E, Klessig, D.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Y7I
 
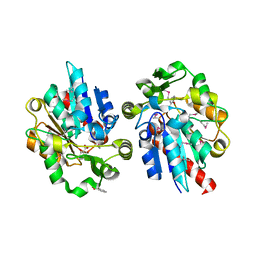 | | Structural and biochemical studies identify tobacco SABP2 as a methylsalicylate esterase and further implicate it in plant innate immunity, Northeast Structural Genomics Target AR2241 | | Descriptor: | 2-HYDROXYBENZOIC ACID, salicylic acid-binding protein 2 | | Authors: | Forouhar, F, Yang, Y, Kumar, D, Chen, Y, Fridman, E, Park, S.W, Chiang, Y, Acton, T.B, Montelione, G.T, Pichersky, E, Klessig, D.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1RZW
 
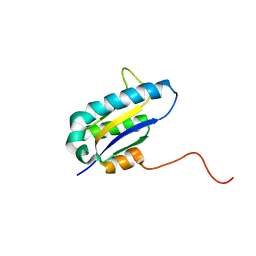 | | The Solution Structure of the Archaeglobus fulgidis protein AF2095. Northeast Structural Genomics Consortium target GR4 | | Descriptor: | Protein AF2095(GR4) | | Authors: | Powers, R, Acton, T.B, Huang, Y.J, Liu, J, Ma, L, Rost, B, Chiang, Y, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-29 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes
Protein Sci., 14, 2005
|
|
5KJS
 
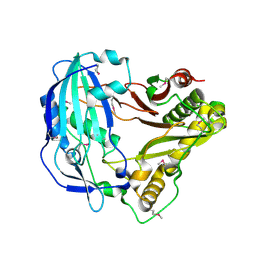 | | Crystal Structure of Arabidopsis thaliana HCT | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJV
 
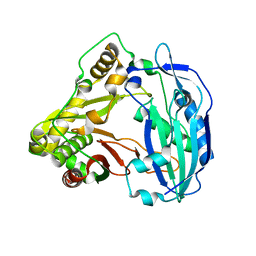 | | Crystal structure of Coleus blumei HCT | | Descriptor: | Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJT
 
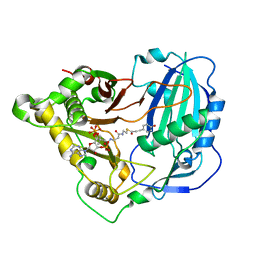 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroyl-CoA | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase, p-coumaroyl-CoA | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJW
 
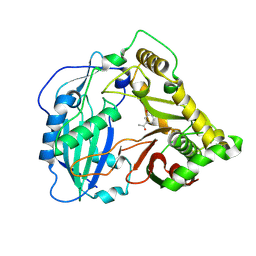 | | Crystal structure of Coleus blumei HCT in complex with 3-hydroxyacetophenone | | Descriptor: | 1-(3-hydroxyphenyl)ethanone, Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJU
 
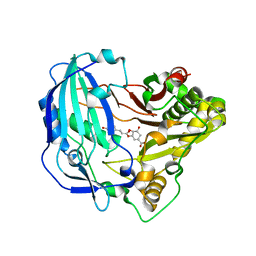 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroylshikimate | | Descriptor: | (3~{R},4~{S},5~{R})-3-[(~{E})-3-(4-hydroxyphenyl)prop-2-enoyl]oxy-4,5-bis(oxidanyl)cyclohexene-1-carboxylic acid, Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
1L7Y
 
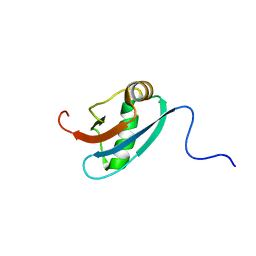 | | Solution NMR Structure of C. elegans Protein ZK652.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR41. | | Descriptor: | HYPOTHETICAL PROTEIN ZK652.3 | | Authors: | Cort, J.R, Chiang, Y, Zheng, D, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-18 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of conserved eukaryotic protein ZK652.3 from C. elegans: a ubiquitin-like fold.
Proteins, 48, 2002
|
|
6EEM
 
 | | Crystal structure of Papaver somniferum tyrosine decarboxylase in complex with L-tyrosine | | Descriptor: | N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-tyrosine, SULFATE ION, TYROSINE, ... | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.61000657 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1SOU
 
 | | NMR structure of Aquifex aeolicus 5,10-methenyltetrahydrofolate synthetase: Northeast Structural Genomics Consortium Target QR46 | | Descriptor: | 5,10-methenyltetrahydrofolate synthetase | | Authors: | Cort, J.R, Chiang, Y, Acton, T, Wu, M, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-15 | | Release date: | 2004-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Aquifex aeolicus 5,10-methenyltetrahydrofolate synthetase: Northeast Structural Genomics Consortium Target QR46
To be Published
|
|
1SC0
 
 | | X-ray Structure of YB61_HAEIN Northeast Structural Genomics Consortium Target IR63 | | Descriptor: | Hypothetical protein HI1161 | | Authors: | Kuzin, A.P, Lee, I, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-11 | | Release date: | 2004-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray Structure of YB61_HAEIN Northeast Structural Genomics Consortium Target IR63.
To be Published
|
|
1SBK
 
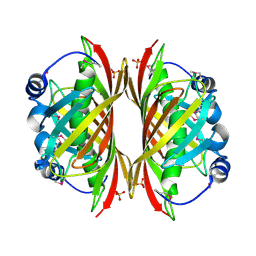 | | X-RAY STRUCTURE OF YDII_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER29. | | Descriptor: | Hypothetical protein ydiI, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Vorobiev, S.M, Lee, I, Forouhar, F, Ma, L, Chiang, Y, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-10 | | Release date: | 2004-02-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Structure of YDII_ECOLI Northeast Structural Genomics Consortium Target ER29
To be Published
|
|
1TIY
 
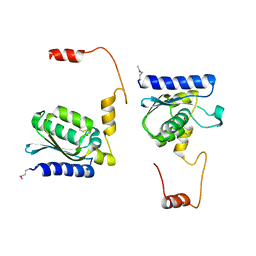 | | X-RAY STRUCTURE OF GUANINE DEAMINASE FROM BACILLUS SUBTILIS NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR160 | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Kuzin, A.P, Vorobiev, S, Edstrom, W, Forouhar, F, Acton, T, Shastry, R, Ma, L.-C, Chiang, Y.-W, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-RAY STRUCTURE OF GUANINE DEAMINASE FROM BACILLUS SUBTILIS
NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR160
To be published
|
|
2KCX
 
 | |
1NI7
 
 | | NORTHEAST STRUCTURAL GENOMIC CONSORTIUM TARGET ER75 | | Descriptor: | Hypothetical protein ygdK | | Authors: | Liu, G, Chiang, Y, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-21 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-quality homology models derived from NMR and X-ray structures of E. coli proteins YgdK and Suf E suggest that all members of the YgdK/Suf E protein family are enhancers of cysteine desulfurases.
Protein Sci., 14, 2005
|
|
