3A8G
 
 | | Crystal structure of Nitrile Hydratase mutant S113A complexed with Trimethylacetonitrile | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
3A8M
 
 | | Crystal structure of Nitrile Hydratase mutant Y72F complexed with Trimethylacetonitrile | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
2RSG
 
 | | Solution structure of the CERT PH domain | | Descriptor: | Collagen type IV alpha-3-binding protein | | Authors: | Sugiki, T, Takeuchi, K, Tokunaga, Y, Kumagai, K, Kawano, M, Nishijima, M, Hanada, K, Takahashi, H, Shimada, I. | | Deposit date: | 2012-02-25 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the Golgi association by the pleckstrin homology domain of the ceramide trafficking protein (CERT)
J.Biol.Chem., 287, 2012
|
|
1BCO
 
 | | BACTERIOPHAGE MU TRANSPOSASE CORE DOMAIN | | Descriptor: | BACTERIOPHAGE MU TRANSPOSASE | | Authors: | Rice, P.A, Mizuuchi, K. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration.
Cell(Cambridge,Mass.), 82, 1995
|
|
1JIB
 
 | | Complex of Alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with Maltotetraose Based on a Crystal Soaked with Maltohexaose. | | Descriptor: | NEOPULLULANASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2001-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1JO8
 
 | | Structural analysis of the yeast actin binding protein Abp1 SH3 domain | | Descriptor: | ACTIN BINDING PROTEIN, SULFATE ION | | Authors: | Fazi, B, Cope, M.J, Douangamath, A, Ferracuti, S, Schirwitz, K, Zucconi, A, Drubin, D.G, Wilmanns, M, Cesareni, G, Castagnoli, L. | | Deposit date: | 2001-07-27 | | Release date: | 2002-03-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unusual binding properties of the SH3 domain of the yeast actin-binding protein Abp1: structural and functional analysis.
J.Biol.Chem., 277, 2002
|
|
3WN8
 
 | | Crystal Structure of Collagen-Model Peptide, (POG)3-PRG-(POG)4 | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Haga, M, Noguchi, K, Tanaka, T. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Preferred side-chain conformation of arginine residues in a triple-helical structure.
Biopolymers, 101, 2014
|
|
1PRV
 
 | | PURINE REPRESSOR DNA-BINDING DOMAIN DNA BINDING | | Descriptor: | PURINE REPRESSOR | | Authors: | Nagadoi, A, Morikawa, S, Nakamura, H, Enari, M, Kobayashi, K, Yamamoto, H, Sampei, G, Mizobuchi, K, Schumacher, M.A, Brennan, R.G, Nishimura, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain.
Structure, 3, 1995
|
|
1PRU
 
 | | PURINE REPRESSOR DNA-BINDING DOMAIN DNA BINDING | | Descriptor: | PURINE REPRESSOR | | Authors: | Nagadoi, A, Morikawa, S, Nakamura, H, Enari, M, Kobayashi, K, Yamamoto, H, Sampei, G, Mizobuchi, K, Schumacher, M.A, Brennan, R.G, Nishimura, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain.
Structure, 3, 1995
|
|
1QTR
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE PROLYL AMINOPEPTIDASE FROM SERRATIA MARCESCENS | | Descriptor: | PROLYL AMINOPEPTIDASE | | Authors: | Yoshimoto, T, Kabashima, T, Uchikawa, K, Inoue, T, Tanaka, N. | | Deposit date: | 1999-06-28 | | Release date: | 1999-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of prolyl aminopeptidase from Serratia marcescens.
J.Biochem.(Tokyo), 126, 1999
|
|
1IYS
 
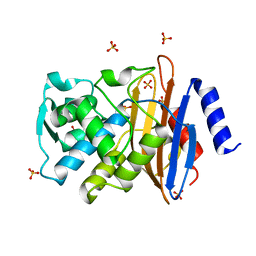 | | Crystal Structure of Class A beta-Lactamase Toho-1 | | Descriptor: | BETA-LACTAMASE TOHO-1, SULFATE ION | | Authors: | Ibuka, A.S, Ishii, Y, Yamaguchi, K, Matsuzawa, H, Sakai, H. | | Deposit date: | 2002-09-06 | | Release date: | 2003-10-14 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Extended-Spectrum beta-Lactamase Toho-1: Insights into the Molecular Mechanism for
Catalytic Reaction and Substrate Specificity Expansion
Biochemistry, 42, 2003
|
|
1RKR
 
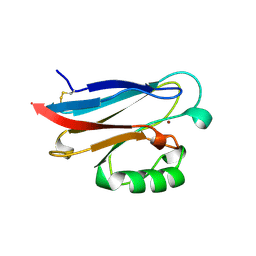 | | CRYSTAL STRUCTURE OF AZURIN-I FROM ALCALIGENES XYLOSOXIDANS NCIMB 11015 | | Descriptor: | AZURIN-I, COPPER (II) ION | | Authors: | Li, C, Inoue, T, Gotowda, M, Suzuki, S, Yamaguchi, K, Kataoka, K, Kai, Y. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of azurin I from the denitrifying bacterium Alcaligenes xylosoxidans NCIMB 11015 at 2.45 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
7UXX
 
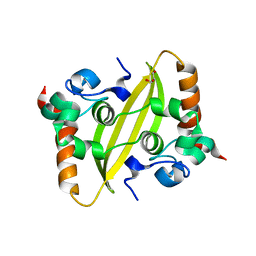 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain | | Descriptor: | ACETATE ION, GLYCEROL, Nucleoprotein | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
7UXZ
 
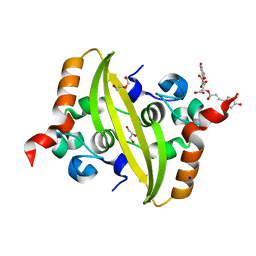 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain complexed with Chicoric acid | | Descriptor: | (2R,3R)-2,3-bis{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}butanedioic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
3B0C
 
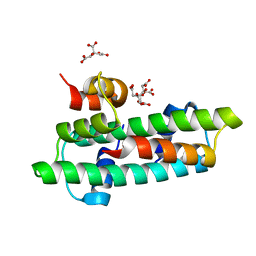 | | Crystal structure of the chicken CENP-T histone fold/CENP-W complex, crystal form I | | Descriptor: | CITRIC ACID, Centromere protein T, Centromere protein W | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-06-08 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
3B0B
 
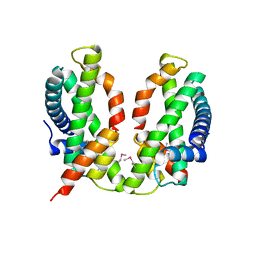 | | Crystal structure of the chicken CENP-S/CENP-X complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-06-08 | | Release date: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
7N30
 
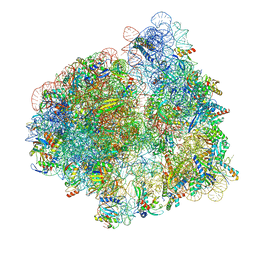 | | Elongating 70S ribosome complex in a hybrid-H2* pre-translocation (PRE-H2*) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-30 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N2C
 
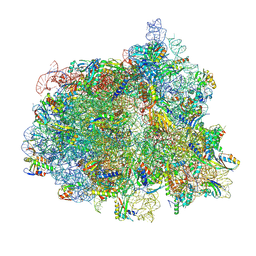 | | Elongating 70S ribosome complex in a fusidic acid-stalled intermediate state of translocation bound to EF-G(GDP) (INT2) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N31
 
 | | Elongating 70S ribosome complex in a post-translocation (POST) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
1L4V
 
 | | SOLUTION STRUCTURE OF SAPECIN | | Descriptor: | Sapecin | | Authors: | Hanzawa, H, Iwai, H, Takeuchi, K, Kuzuhara, T, Komano, H, Kohda, D, Inagaki, F, Natori, S, Arata, Y, Shimada, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-03-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | 1H nuclear magnetic resonance study of the solution conformation of an antibacterial protein, sapecin.
FEBS Lett., 269, 1990
|
|
7N2U
 
 | | Elongating 70S ribosome complex in a hybrid-H1 pre-translocation (PRE-H1) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N2V
 
 | | Elongating 70S ribosome complex in a spectinomycin-stalled intermediate state of translocation bound to EF-G in an active, GTP conformation (INT1) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
2RQZ
 
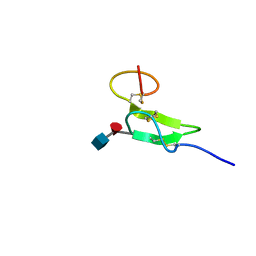 | | Structure of sugar modified epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose, Neurogenic locus notch homolog protein 1 | | Authors: | Shimizu, K, Fujitani, N, Hosoguchi, K, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
3VQJ
 
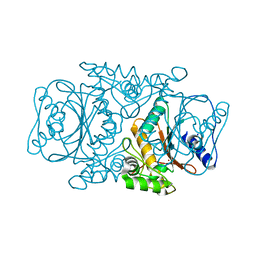 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, ZINC ION | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-03-24 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
3VRK
 
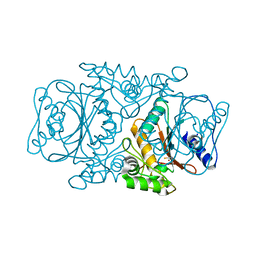 | | Crystal Structutre of Thiobacillus thioparus THI115 Carbonyl Sulfide Hydrolase / Thiocyanate complex | | Descriptor: | Carbonyl sulfide hydrolase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Katayama, Y, Noguchi, K, Ogawa, T, Ohtaki, A, Odaka, M, Yohda, M. | | Deposit date: | 2012-04-11 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
J.Am.Chem.Soc., 135, 2013
|
|
