8G6I
 
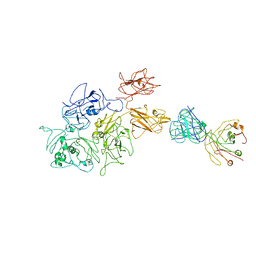 | | Coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor VIII chimera from human and pig, NB33 heavy chain, ... | | Authors: | Childers, K.C, Davulcu, O, Haynes, R.M, Lollar, P, Doering, C.B, Coxon, C.H, Spiegel, P.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor.
Blood, 142, 2023
|
|
4YS2
 
 | | RCK domain with CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Na+/H+ antiporter-like protein | | Authors: | Chin, K.H, Chou, S.H. | | Deposit date: | 2015-03-16 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | RCK domain with CDA
To Be Published
|
|
6WNM
 
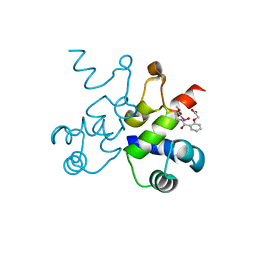 | | The structure of Pf4r from a superinfective isolate of the filamentous phage Pf4 of Pseudomonas aeruginosa PA01 | | Descriptor: | Pf4r, [(2R)-3-{[2-(carboxymethoxy)benzene-1-carbonyl]amino}-2-methoxypropyl](hydroxy)mercury | | Authors: | Michie, K.A, Norrian, P, Duggin, I.G, McDougald, D, Rice, S.A. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The Repressor C Protein, Pf4r, Controls Superinfection of Pseudomonas aeruginosa PAO1 by the Pf4 Filamentous Phage and Regulates Host Gene Expression.
Viruses, 13, 2021
|
|
6WPZ
 
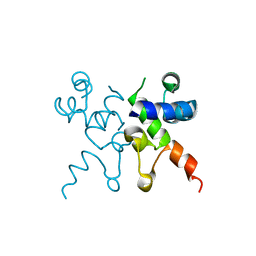 | | The structure of Pf4r from a superinfective isolate of the filamentous phage Pf4 of Pseudomonas aeruginosa PA01 | | Descriptor: | CHLORIDE ION, Pf4r | | Authors: | Michie, K.A, Norrian, P, Duggin, I.G, McDougald, D, Rice, S.A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | The Repressor C Protein, Pf4r, Controls Superinfection of Pseudomonas aeruginosa PAO1 by the Pf4 Filamentous Phage and Regulates Host Gene Expression.
Viruses, 13, 2021
|
|
6LP1
 
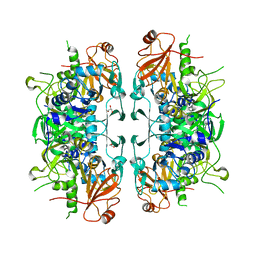 | | Crystal structure of acetate:succinate CoA transferase (ASCT) from Trypanosoma brucei. | | Descriptor: | CALCIUM ION, GLYCEROL, Succinyl-CoA:3-ketoacid-coenzyme A transferase | | Authors: | Mochizuki, K, Inaoka, D.K, Shiba, T, Fukuda, K, Kurasawa, H, Mazet, M, Millerioux, Y, Bringaud, F, Boshart, M, Balogun, E.O, Harada, S, Hirayama, K, Kita, K. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The ASCT/SCS cycle fuels mitochondrial ATP and acetate production in Trypanosoma brucei.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
5L0H
 
 | |
5L0I
 
 | |
5L0J
 
 | |
5NYM
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in reduced state | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5NYK
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in oxidized state | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5B5L
 
 | | Crystal structure of acetyl esterase mutant S10A with acetate ion | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Uechi, K, Kamachi, S, Akita, H, Mine, S, Watanabe, M. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | crystal structure of acetyl esterase mutant S10A with acetate ion
To Be Published
|
|
5NYO
 
 | | Crystal structure of an atypical poplar thioredoxin-like2.1 variant in dimeric form | | Descriptor: | SULFATE ION, Thioredoxin-like protein 2.1 | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
2J6I
 
 | |
5NYN
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in complex with gluathione | | Descriptor: | GLUTATHIONE, SULFATE ION, Thioredoxin-like protein 2.1 | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5NYL
 
 | |
6CDS
 
 | | Human neurofibromin 2/merlin/schwannomin residues 1-339 in complex with PIP2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Merlin, ... | | Authors: | Chinthalapudi, K, Sharff, A.J, Bricogne, G, Izard, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Lipid binding promotes the open conformation and tumor-suppressive activity of neurofibromin 2.
Nat Commun, 9, 2018
|
|
1OVF
 
 | | NMR Structure of ActD/5'-CCGTTTTGTGG-3' Complex | | Descriptor: | (5'-D(*CP*CP*GP*TP*TP*TP*TP*GP*TP*GP*G)-3'), ACTINOMYCIN D | | Authors: | Chin, K.-H, Chou, S.-H, Chen, F.-M. | | Deposit date: | 2003-03-26 | | Release date: | 2003-05-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Actd-5'-Ccgtt(3)Gtgg-3' Complex: Drug Interaction with Tandem G.T Mismatches and Hairpin Loop Backbone.
Nucleic Acids Res., 31, 2003
|
|
8CAS
 
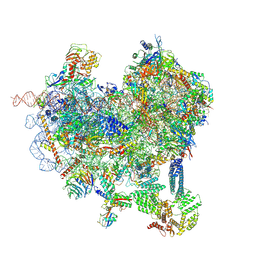 | | Cryo-EM structure of native Otu2-bound ubiquitinated 48S initiation complex (partial) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8CBJ
 
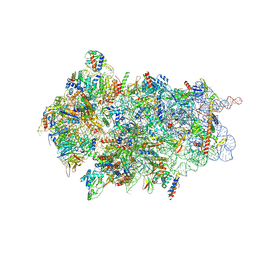 | | Cryo-EM structure of Otu2-bound cytoplasmic pre-40S ribosome biogenesis complex | | Descriptor: | 20S pre-ribosomal RNA, 20S-pre-rRNA D-site endonuclease NOB1, 40S ribosomal protein S0-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
4KBY
 
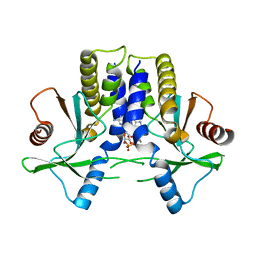 | | mSTING/c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Stimulator of interferon genes protein | | Authors: | Chin, K.H, Su, Y.C, Tu, J.L, Chou, S.H. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2JOG
 
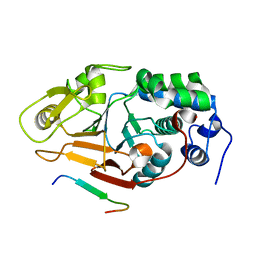 | | Structure of the calcineurin-NFAT complex | | Descriptor: | Calmodulin-dependent calcineurin A subunit alpha isoform, NFAT | | Authors: | Takeuchi, K, Roehrl, M.H, Sun, Z.Y, Wagner, G. | | Deposit date: | 2007-03-10 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the Calcineurin-NFAT Complex: Defining a T Cell Activation Switch Using Solution NMR and Crystal Coordinates.
Structure, 15, 2007
|
|
2JXB
 
 | | Structure of CD3epsilon-Nck2 first SH3 domain complex | | Descriptor: | T-cell surface glycoprotein CD3 epsilon chain, Cytoplasmic protein NCK2 | | Authors: | Takeuchi, K, Yang, H, Ng, E, Park, S, Sun, Z.J, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-11-09 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional evidence that Nck interaction with CD3epsilon regulates T-cell receptor activity.
J.Mol.Biol., 380, 2008
|
|
4KC0
 
 | | mSTING | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Chin, K.H, Su, Y.C, Tu, J.L, Chou, S.H. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5H5O
 
 | |
7T8S
 
 | | Light Harvesting complex phycoerythrin PE 566, from the cryptophyte Cryptomonas pyrenoidifera | | Descriptor: | Bilin 584 (doubly linked), Bilin 584 (single linked), Bilin 618 (single linked), ... | | Authors: | Michie, K.A, Harrop, S.J, Rathbone, H.W, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2021-12-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structures reveal the origin of spectral variation in cryptophyte light harvesting antenna proteins.
Protein Sci., 32, 2023
|
|
