5VPJ
 
 | | The crystal structure of a thioesteras from Actinomadura verrucosospora. | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, Thioesterase | | Authors: | Tan, K, Joachimiak, G, Endres, M, Phillips Jr, G.N, Joachmiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-05-05 | | Release date: | 2017-07-19 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of a thioesteras from Actinomadura verrucosospora.
To Be Published
|
|
4E5U
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with thymidine monophosphate. | | Descriptor: | CALCIUM ION, THYMIDINE-5'-PHOSPHATE, Thymidylate kinase | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-03-14 | | Release date: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with thymidine monophosphate.
To be Published
|
|
4GMD
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with AZT Monophosphate | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-08-15 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with AZT Monophosphate
To be Published
|
|
4MPT
 
 | | Crystal Structure of Periplasmic binding Protein Type 1 from Bordetella pertussis Tohama I | | Descriptor: | ACETIC ACID, Putative leu/ile/val-binding protein, SODIUM ION | | Authors: | Kim, Y, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-13 | | Release date: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Periplasmic binding Protein Type 1 from Bordetella pertussis Tohama I
To be Published
|
|
4P98
 
 | |
4J00
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 24mer DNA target | | Descriptor: | DNA (5'-D(*TP*GP*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*CP*C)-3'), MAGNESIUM ION, Transcription Factor HetR | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1R7L
 
 | | 2.0 A Crystal Structure of a Phage Protein from Bacillus cereus ATCC 14579 | | Descriptor: | Phage protein | | Authors: | Zhang, R, Joachimiak, G, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of phage protein BC1872 from Bacillus cereus, a singleton with new fold
Proteins, 64, 2006
|
|
4ESH
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with deoxythymidine. | | Descriptor: | THYMIDINE, Thymidylate kinase | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with deoxythymidine.
To be Published
|
|
8U01
 
 | | Crystal Structure of the Glycoside Hydrolase Family 2 TIM Barrel-domain Containing Protein from Phocaeicola plebeius | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2023-08-28 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Glycoside Hydrolase Family 2 TIM Barrel-domain
Containing Protein from Phocaeicola plebeius
To Be Published
|
|
1N5Q
 
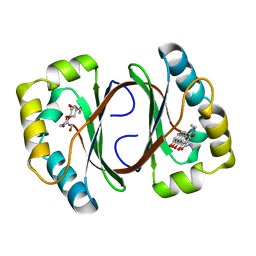 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with dehydrated Sancycline | | Descriptor: | 4-DIMETHYLAMINO-1,10,11,12-TETRAHYDROXY-3-OXO-3,4,4A,5-TETRAHYDRO-NAPHTHACENE-2-CARBOXYLIC ACID AMIDE, ActaVA-Orf6 monooxygenase, HEXAETHYLENE GLYCOL | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5S
 
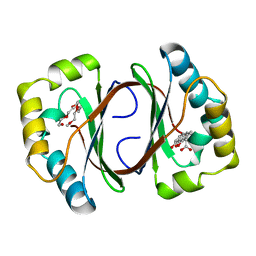 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Acetyl Dithranol | | Descriptor: | (1,8-DIHYDROXY-9-OXO-9,10-DIHYDRO-ANTHRACEN-2-YL)-ACETIC ACID, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5T
 
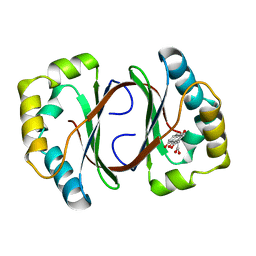 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Oxidized Acetyl Dithranol | | Descriptor: | (1,8-DIHYDROXY-9,10-DIOXO-9,10-DIHYDRO-ANTHRACEN-2-YL)-ACETIC ACID, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, G Kendrew, S, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5V
 
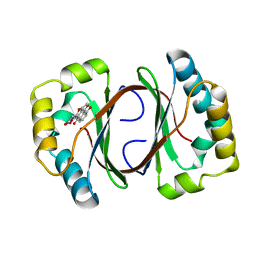 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Nanaomycin D | | Descriptor: | 7-HYDROXY-5-METHYL-3,3A,5,11B-TETRAHYDRO-1,4-DIOXA-CYCLOPENTA[A]ANTHRACENE-2,6,11-TRIONE, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
4KQC
 
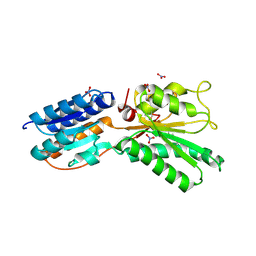 | | ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii | | Descriptor: | NITRATE ION, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Osipiuk, J, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-14 | | Release date: | 2013-05-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii
To be Published
|
|
4IZZ
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 21mer DNA target | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*C)-3'), SULFATE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1SQE
 
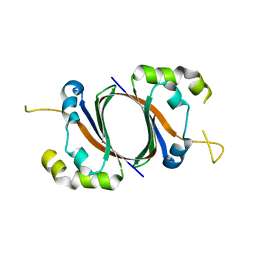 | | 1.5A Crystal Structure Of the protein PG130 from Staphylococcus aureus, Structural genomics | | Descriptor: | hypothetical protein PG130 | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases
J.Biol.Chem., 280, 2005
|
|
3FGG
 
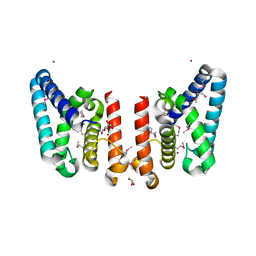 | | Crystal Structure of Putative ECF-type Sigma Factor Negative Effector from Bacillus cereus | | Descriptor: | GLYCEROL, ZINC ION, uncharacterized protein BCE2196 | | Authors: | Kim, Y, Nocek, B, Maltseva, N, Joachimiak, G, Du, J, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-05 | | Release date: | 2009-01-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Putative ECF-type Sigma Factor Negative Effector from Bacillus cereus
To be Published
|
|
1NG5
 
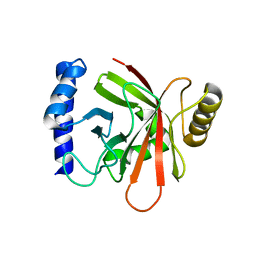 | |
3GE2
 
 | | Crystal structure of putative lipoprotein SP_0198 from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein, ... | | Authors: | Kim, Y, Zhang, R, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Putative Lipoprotein SP_0198 from Streptococcus pneumoniae
To be Published
|
|
5E2F
 
 | | Crystal Structure of Beta-lactamase class D from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase YbxI, CALCIUM ION | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Beta-lactamase class D from Bacillus subtilis
To Be Published
|
|
5E2G
 
 | | Crystal Structure of D-alanine Carboxypeptidase AmpC from Burkholderia cenocepacia | | Descriptor: | ACETIC ACID, Beta-lactamase, THIOCYANATE ION | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal Structure of D-alanine Carboxypeptidase AmpC from Burkholderia cenocepacia
To Be Published
|
|
2QM1
 
 | | Crystal structure of glucokinase from Enterococcus faecalis | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glucokinase, ... | | Authors: | Kim, Y, Joachimiak, G, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of Glucokinase from Enterococcus faecalis.
To be Published
|
|
2QQZ
 
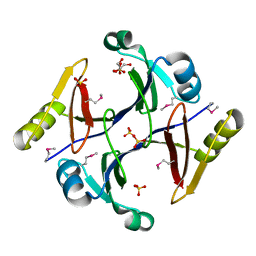 | | Crystal structure of putative glyoxalase family protein from Bacillus anthracis | | Descriptor: | GLYCEROL, Glyoxalase family protein, putative, ... | | Authors: | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Putative Glyoxalase Family Protein from Bacillus anthracis.
To be Published
|
|
2IZX
 
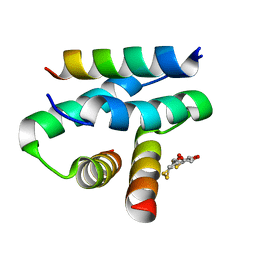 | | Molecular Basis of AKAP Specificity for PKA Regulatory Subunits | | Descriptor: | AKAP-IS, CAMP-DEPENDENT PROTEIN KINASE TYPE II-ALPHA REGULATORY SUBUNIT, DITHIANE DIOL | | Authors: | Gold, M.G, Lygren, B, Dokurno, P, Hoshi, N, McConnachie, G, Tasken, K, Carlson, C.R, Scott, J.D, Barford, D. | | Deposit date: | 2006-07-27 | | Release date: | 2006-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular Basis of Akap Specificity for Pka Regulatory Subunits.
Mol.Cell, 24, 2006
|
|
2IZY
 
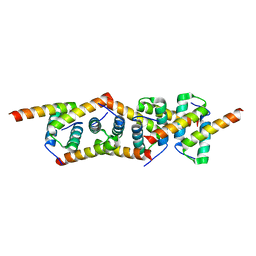 | | Molecular Basis of AKAP Specificity for PKA Regulatory Subunits | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE REGULATORY SUBUNIT II | | Authors: | Gold, M.G, Lygren, B, Dokurno, P, Hoshi, N, McConnachie, G, Tasken, K, Carlson, C.R, Scott, J.D, Barford, D. | | Deposit date: | 2006-07-27 | | Release date: | 2006-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis of Akap Specificity for Pka Regululatory Subunits
Mol.Cell, 24, 2006
|
|
