1A41
 
 | | TYPE 1-TOPOISOMERASE CATALYTIC FRAGMENT FROM VACCINIA VIRUS | | Descriptor: | SULFATE ION, TOPOISOMERASE I | | Authors: | Cheng, C, Kussie, P, Pavletich, N, Shuman, S. | | Deposit date: | 1998-02-10 | | Release date: | 1999-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases.
Cell(Cambridge,Mass.), 92, 1998
|
|
3UT7
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3UT8
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | IODIDE ION, Putative uncharacterized protein | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3UT4
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3JZI
 
 | | Crystal structure of biotin carboxylase from E. Coli in complex with benzimidazole series | | Descriptor: | 7-amino-2-[(2-chlorobenzyl)amino]-1-{[(1S,2S)-2-hydroxycycloheptyl]methyl}-1H-benzimidazole-5-carboxamide, Biotin carboxylase | | Authors: | Cheng, C, Shipps, G.W, Yang, Z, Sun, B, Kawahata, N, Soucy, K, Soriano, A, Orth, P, Xiao, L, Mann, P, Black, T. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and optimization of antibacterial AccC inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5ZVB
 
 | | APOBEC3F Chimeric Catalytic Domain in Complex with DNA(dT9) | | Descriptor: | APEBEC3F/ssDNA-T9, CACODYLATE ION, DNA (5'-D(*AP*TP*TP*TP*TP*CP*AP*AP*T)-3'), ... | | Authors: | Cheng, C, Zhang, T.L, Wang, C.X, Lan, W.X, Ding, J.P, Cao, C.Y. | | Deposit date: | 2018-05-09 | | Release date: | 2018-11-21 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Cytidine Deaminase Human APOBEC3F Chimeric Catalytic Domain in Complex with DNA
Chin.J.Chem., 36, 2018
|
|
5ZVA
 
 | | APOBEC3F Chimeric Catalytic Domain in Complex with DNA(dC9) | | Descriptor: | APEBEC3F/ssDNA-C9, CACODYLATE ION, DNA (5'-D(*AP*TP*TP*TP*TP*CP*AP*AP*CP*T)-3'), ... | | Authors: | Cheng, C, Zhang, T.L, Wang, C.X, Lan, W.X, Ding, J.P, Cao, C.Y. | | Deposit date: | 2018-05-09 | | Release date: | 2018-11-21 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Cytidine Deaminase Human APOBEC3F Chimeric Catalytic Domain in Complex with DNA
Chin.J.Chem., 36, 2018
|
|
6J02
 
 | | Crystal structure of the SRCR domain of mouse SCARA1 | | Descriptor: | CALCIUM ION, Macrophage scavenger receptor types I and II | | Authors: | Cheng, C, Hu, Z. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | The scavenger receptor SCARA1 (CD204) recognizes dead cells through spectrin.
J.Biol.Chem., 294, 2019
|
|
7DPX
 
 | |
2WRY
 
 | | Crystal structure of chicken cytokine interleukin 1 beta | | Descriptor: | INTERLEUKIN-1BETA | | Authors: | Lu, W.S, Cheng, C.S, Lyu, P.C, Lee, L.H, Wang, W.C, Yin, H.S. | | Deposit date: | 2009-09-03 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Functional Comparison of Cytokine Interleukin-1 Beta from Chicken and Human.
Mol.Immunol., 48, 2011
|
|
5E26
 
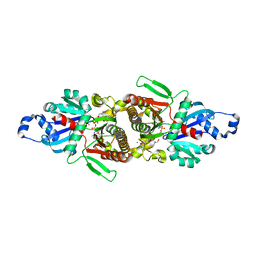 | | Crystal structure of human PANK2: the catalytic core domain in complex with pantothenate and adenosine diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | DONG, A, LOPPNAU, P, RAVICHANDRAN, M, CHENG, C, TEMPEL, W, SEITOVA, A, HUTCHINSON, A, HONG, B.S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of human PANK2: the catalytic core domain in complex with pantothenate and adenosine diphosphate
to be published
|
|
3OTW
 
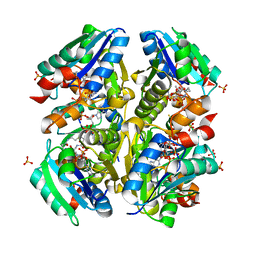 | | Structural and Functional Studies of Helicobacter pylori Wild-Type and Mutated Proteins Phosphopantetheine adenylyltransferase | | Descriptor: | COENZYME A, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Yin, H.S, Cheng, C.S, Chen, C.G, Luo, Y.C, Chen, W.T, Cheng, S.Y. | | Deposit date: | 2010-09-14 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Studies of Helicobacter pylori Wild-Type and Mutated Proteins Phosphopantetheine adenylyltransferase
To be Published
|
|
1TI5
 
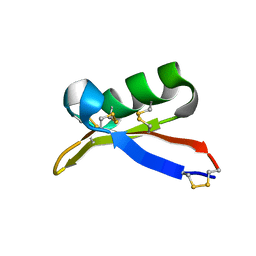 | | Solution structure of plant defensin | | Descriptor: | plant defensin | | Authors: | Liu, Y.J, Cheng, C.S, Liu, Y.N, Hsu, M.P, Chen, C.S, Lyu, P.C. | | Deposit date: | 2004-06-02 | | Release date: | 2005-07-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the plant defensin VrD1 from mung bean and its possible role in insecticidal activity against bruchids.
Proteins, 63, 2006
|
|
7QO4
 
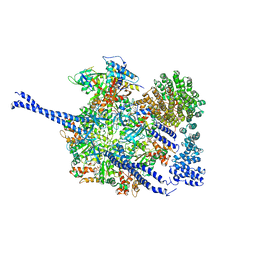 | | 26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn1, Ubp6, and UbVS) | | Descriptor: | 26S proteasome regulatory subunit RPN1, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
2O4X
 
 | | Crystal structure of human P100 tudor domain | | Descriptor: | Staphylococcal nuclease domain-containing protein 1 | | Authors: | Shaw, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Rao, Z, Wang, B.C, Liu, Z.J. | | Deposit date: | 2006-12-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human P100 tudor domain
To be Published
|
|
1SIY
 
 | | NMR structure of mung bean non-specific lipid transfer protein 1 | | Descriptor: | Nonspecific lipid-transfer protein 1 | | Authors: | Lin, K.F, Liu, Y.N, Hsu, S.T.D, Samuel, D, Cheng, C.S, Bonvin, A.M.J.J, Lyu, P.C. | | Deposit date: | 2004-03-02 | | Release date: | 2005-04-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Characterization and Structural Analyses of Nonspecific Lipid Transfer Protein 1 from Mung Bean
Biochemistry, 44, 2005
|
|
3ECR
 
 | | Structure of human porphobilinogen deaminase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Song, G, Li, Y, Cheng, C, Zhao, Y, Gao, A, Zhang, R, Joachimiak, A, Shaw, N, Liu, Z.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structural insight into acute intermittent porphyria.
Faseb J., 23, 2009
|
|
2HY6
 
 | | A seven-helix coiled coil | | Descriptor: | General control protein GCN4, HEXANE-1,6-DIOL | | Authors: | Liu, J, Zheng, Q, Deng, Y, Cheng, C.S, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A seven-helix coiled coil.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2L95
 
 | | Solution Structure of Cytotoxic T-Lymphocyte Antigent-2(Ctla protein), Crammer at pH 6.0 | | Descriptor: | Crammer | | Authors: | Tseng, T.S, Cheng, C.S, Liu, Y.N, Lyu, P.C. | | Deposit date: | 2011-01-31 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A molten globule-to-ordered structure transition of Drosophila melanogaster crammer is required for its ability to inhibit cathepsin.
Biochem.J., 442, 2012
|
|
2HQE
 
 | | Crystal structure of human P100 Tudor domain: Large fragment | | Descriptor: | P100 Co-activator tudor domain | | Authors: | Shah, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Liu, Z.J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a large fragment of the Human P100 Tudor Domain
To be Published
|
|
2HQ1
 
 | | Crystal Structure of ORF 1438 a putative Glucose/ribitol dehydrogenase from Clostridium thermocellum | | Descriptor: | Glucose/ribitol dehydrogenase | | Authors: | Southeast Collaboratory for Structural Genomics (SECSG), Li, Y, Shaw, N, Xu, H, Cheng, C, Chen, L, Liu, Z.J, Rose, J.P, Wang, B.C. | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of ORF 1438 a putative Glucose/ ribitol dehydrogenase from Clostridium thermocellum
To be Published
|
|
3NV7
 
 | |
3HY6
 
 | | Structure of human MTHFS with ADP | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3HY3
 
 | | Structure of human MTHFS with 10-formyltetrahydrofolate | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, N-({4-[{[(2R,4S,4aR,6S,8aS)-2-amino-4-hydroxydecahydropteridin-6-yl]methyl}(formyl)amino]phenyl}carbonyl)-D-glutamic acid, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3HXT
 
 | | Structure of human MTHFS | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, NICKEL (II) ION | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Zhang, R, Joachimiak, A, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
