4RWT
 
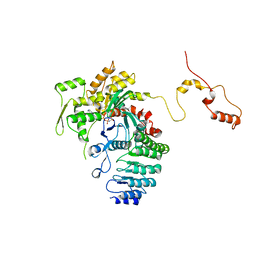 | | Structure of actin-Lmod complex | | Descriptor: | Actin-5C, Leiomodin-2, MAGNESIUM ION, ... | | Authors: | Chen, X, Ni, F, Wang, Q. | | Deposit date: | 2014-12-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Mechanisms of leiomodin 2-mediated regulation of actin filament in muscle cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2NBV
 
 | |
4XKC
 
 | |
4XKB
 
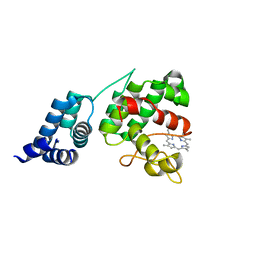 | | Crystal Structure of GENOMES UNCOUPLED 4 (GUN4) in Complex with Deuteroporphyrin IX | | Descriptor: | 3,3'-(3,7,12,17-tetramethylporphyrin-2,18-diyl)dipropanoic acid, Ycf53-like protein | | Authors: | Chen, X, Pu, H, Liu, L. | | Deposit date: | 2015-01-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal Structures of GUN4 in Complex with Porphyrins.
Mol Plant, 8, 2015
|
|
4ZHJ
 
 | |
2N3T
 
 | |
6AKO
 
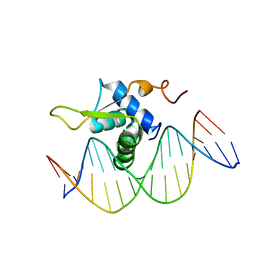 | | Crystal Structure of FOXC2 DBD Bound to DBE2 DNA | | Descriptor: | DNA (5'-D(CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Forkhead box protein C2, ... | | Authors: | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
6AKP
 
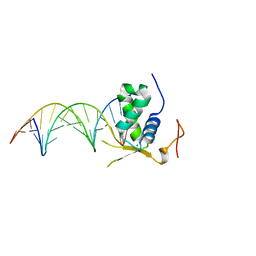 | | Crystal Structural of FOXC2 DNA binding domain bound to PC promoter | | Descriptor: | DNA (5'-D(AP*CP*AP*CP*AP*AP*AP*TP*AP*TP*TP*TP*GP*TP*GP*T)-3'), Forkhead box protein C2, MAGNESIUM ION | | Authors: | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
2N3W
 
 | |
2N3U
 
 | |
8YZ5
 
 | | SARS-CoV-2 Delta Spike in complex with Fab of JE-5C | | Descriptor: | Fab heavy chain of JE-5C, Fab light chain of JE-5C, Spike glycoprotein | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2024-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YZ6
 
 | | SARS-CoV-2 Spike (BA.1) in complex with Fab of JH-8B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain of JH-8B, ... | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2024-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8Y22
 
 | | FGFR1 kinase domain with a covalent inhibitor 9g | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, ~{N}-[4-[[4-azanyl-3-(7-methoxy-5-methyl-1-benzothiophen-2-yl)pyrazolo[3,4-d]pyrimidin-1-yl]methyl]phenyl]propanamide | | Authors: | Chen, X.J, Chen, Y.H. | | Deposit date: | 2024-01-25 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Design, synthesis and biological evaluation of 5-amino-1H-pyrazole-4-carboxamide derivatives as pan-FGFR covalent inhibitors.
Eur.J.Med.Chem., 275, 2024
|
|
8XZ7
 
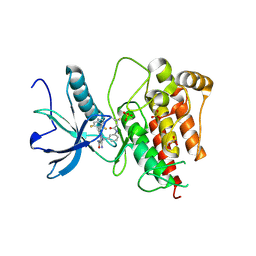 | | FGFR1 kinase domain with a covalent inhibitor 10h | | Descriptor: | 5-azanyl-3-[2-[4,6-bis(fluoranyl)-2-methyl-3~{H}-benzimidazol-5-yl]ethynyl]-1-[[3-(prop-2-enoylamino)phenyl]methyl]pyrazole-4-carboxamide, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Chen, X.J, Chen, Y.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis and biological evaluation of 5-amino-1H-pyrazole-4-carboxamide derivatives as pan-FGFR covalent inhibitors.
Eur.J.Med.Chem., 275, 2024
|
|
2NBU
 
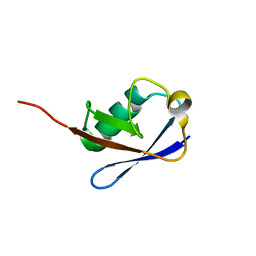 | |
2NBW
 
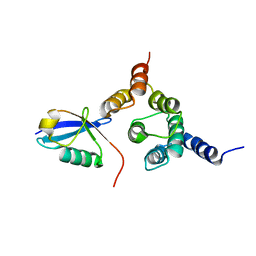 | | Solution structure of the Rpn1 T1 site with the Rad23 UBL domain | | Descriptor: | 26S proteasome regulatory subunit RPN1, UV excision repair protein RAD23 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2016-03-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
1XCA
 
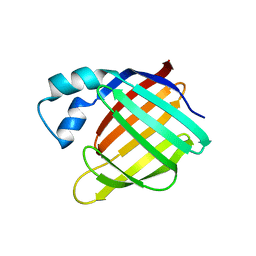 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN TYPE II | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1996-12-31 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of apo-cellular retinoic acid-binding protein type II (R111M) suggests a mechanism of ligand entry.
J.Mol.Biol., 278, 1998
|
|
2N3V
 
 | |
2KQZ
 
 | |
2KR0
 
 | |
3OLJ
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) | | Descriptor: | Ribonucleoside-diphosphate reductase subunit M2, SODIUM ION | | Authors: | Chen, X.H, Xu, Z.J, Chen, B.E, Jiang, H.J, Yang, C.G, Zhu, W.L, Shao, J.M. | | Deposit date: | 2010-08-26 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | hRRM2
To be Published
|
|
5W1X
 
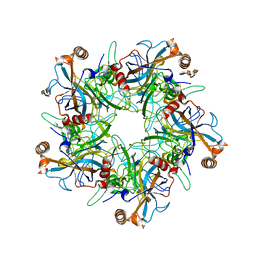 | | Crystal Structure of Humanpapillomavirus18 (HPV18) Capsid L1 Pentamers Bound to Heparin Oligosaccharides | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, ... | | Authors: | Chen, X.S, Dasgupta, J. | | Deposit date: | 2017-06-05 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.374 Å) | | Cite: | Structural basis of oligosaccharide receptor recognition by human papillomavirus.
J. Biol. Chem., 286, 2011
|
|
4TTQ
 
 | |
474D
 
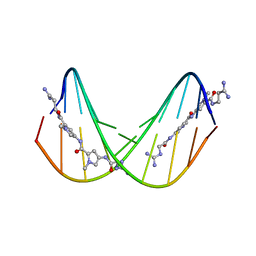 | | A NOVEL END-TO-END BINDING OF TWO NETROPSINS TO THE DNA DECAMER D(CCCCCIIIII)2 | | Descriptor: | DNA (5'-D(*CP*CP*CP*(CBR)P*CP*IP*IP*IP*IP*I)-3'), NETROPSIN | | Authors: | Chen, X, Rao, S.T, Sekar, K, Sundaralingam, M. | | Deposit date: | 1998-01-14 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel end-to-end binding of two netropsins to the DNA decamers d(CCCCCIIIII)2, d(CCCBr5CCIIIII)2and d(CBr5CCCCIIIII)2.
Nucleic Acids Res., 26, 1998
|
|
6K9C
 
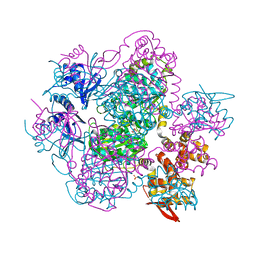 | | The apo structure of NrS-1 C terminal region (305-718) | | Descriptor: | MERCURY (II) ION, Primase, SULFATE ION | | Authors: | Chen, X, Gan, J. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structural studies reveal a ring-shaped architecture of deep-sea vent phage NrS-1 polymerase.
Nucleic Acids Res., 48, 2020
|
|
