8FVI
 
 | | Human APOBEC3H bound to HIV-1 Vif in complex with CBF-beta, ELOB, ELOC, and CUL5 | | Descriptor: | Core-binding factor subunit beta, Cullin 5, DNA dC->dU-editing enzyme APOBEC-3H, ... | | Authors: | Ito, F, Alvarez-Cabrera, A.L, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural basis of HIV-1 Vif-mediated E3 ligase targeting of host APOBEC3H.
Nat Commun, 14, 2023
|
|
7RSL
 
 | | Seipin forms a flexible cage at lipid droplet formation sites | | Descriptor: | Seipin | | Authors: | Arlt, H, Sui, X, Folger, B, Adams, C, Chen, X, Remme, R, Hamprecht, F.A, DiMaio, F, Liao, M, Goodman, J.M, Farese Jr, R.V, Walther, T.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Seipin forms a flexible cage at lipid droplet formation sites.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6K7Z
 
 | | Crystal structure of a GH18 chitinase from Pseudoalteromonas aurantia | | Descriptor: | GH18 chiitnase | | Authors: | Wang, Y.J, Li, P.Y, Cao, H.Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural Insight Into Chitin Degradation and Thermostability of a Novel Endochitinase From the Glycoside Hydrolase Family 18.
Front Microbiol, 10, 2019
|
|
1QET
 
 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*AP*UP*GP*UP*CP*C)-3') | | Authors: | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
5K83
 
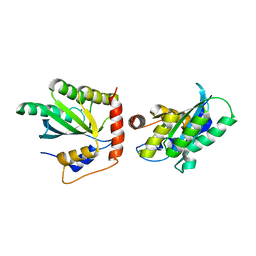 | | Crystal Structure of a Primate APOBEC3G N-Domain, in Complex with ssDNA | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
1QES
 
 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*UP*UP*CP*C)-3') | | Authors: | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
5K81
 
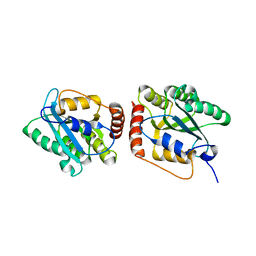 | | Crystal Structure of a Primate APOBEC3G N-Terminal Domain | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
8JOT
 
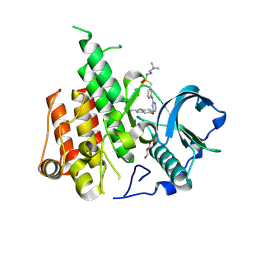 | | Crystal structure of CSF-1R kinase domain with sulfatinib | | Descriptor: | GLYCEROL, Macrophage colony-stimulating factor 1 receptor, N-(2-(dimethylamino)ethyl)-1-(3-((4-((2-methyl-1H-indol-5-yl)oxy)pyrimidin-2-yl)amino)phenyl)methanesulfonamide | | Authors: | Lin, Q.M, Chen, X.J, Chen, Y.H. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis and selectivity of sulfatinib binding to FGFR and CSF-1R.
Commun Chem, 7, 2024
|
|
5K82
 
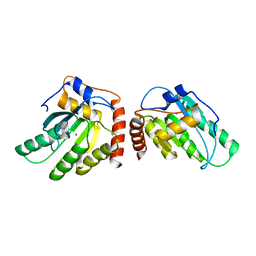 | | Crystal Structure of a Primate APOBEC3G N-Terminal Domain | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
2R5H
 
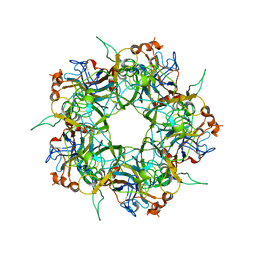 | |
2R5I
 
 | |
7SGL
 
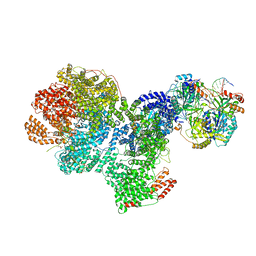 | | DNA-PK complex of DNA end processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, Hairpin_1, ... | | Authors: | Liu, L, Li, J, Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2021-10-06 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7RR5
 
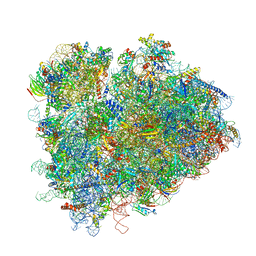 | | Structure of ribosomal complex bound with Rbg1/Tma46 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Zeng, F, Li, X, Pires-Alves, M, Chen, X, Hawk, C.W, Jin, H. | | Deposit date: | 2021-08-09 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells.
Cell Rep, 37, 2021
|
|
1S8K
 
 | | Solution Structure of BmKK4, A Novel Potassium Channel Blocker from Scorpion Buthus martensii Karsch, 25 structures | | Descriptor: | Toxin BmKK4 | | Authors: | Zhang, N, Chen, X, Li, M, Cao, C, Wang, Y, Hu, G, Wu, H. | | Deposit date: | 2004-02-02 | | Release date: | 2005-02-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK4, the first member of subfamily alpha-KTx 17 of scorpion toxins
Biochemistry, 43, 2004
|
|
2Q9Q
 
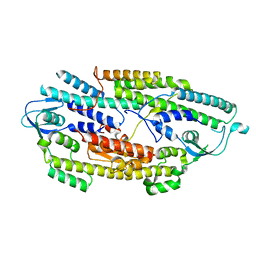 | | The crystal structure of full length human GINS complex | | Descriptor: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, GINS complex subunit 3, ... | | Authors: | Chang, Y.P, Wang, G, Chen, X.S. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of the GINS complex and functional insights into its role in DNA replication.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1GH2
 
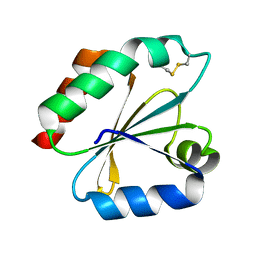 | | Crystal structure of the catalytic domain of a new human thioredoxin-like protein | | Descriptor: | THIOREDOXIN-LIKE PROTEIN | | Authors: | Jin, J, Chen, X, Guo, Q, Yuan, J, Qiang, B, Rao, Z. | | Deposit date: | 2000-11-01 | | Release date: | 2001-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of the catalytic domain of a human thioredoxin-like protein.
Eur.J.Biochem., 269, 2002
|
|
5X0T
 
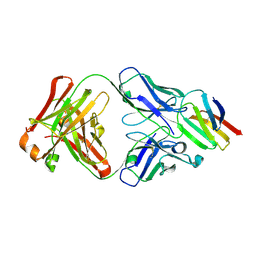 | | Crystal structure of CD147 C2 domain in complex with Fab of its monoclonal antibody 6H8 | | Descriptor: | 6H8 Fab fragment heavy chain, 6H8 Fab fragment light chain, Basigin | | Authors: | Lin, P, Zhang, M.-Y, Ye, S, Chen, X, Yu, X.-L, Zhang, R.-G, Zhu, P, Chen, Z.-N. | | Deposit date: | 2017-01-23 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CD147 C2 domain in complex with Fab of its monoclonal antibody
To Be Published
|
|
1GHQ
 
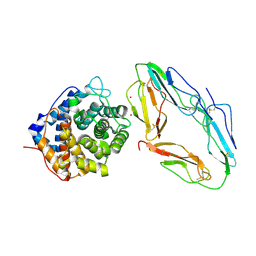 | | CR2-C3D COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, COMPLEMENT C3, CR2/CD121/C3D/EPSTEIN-BARR VIRUS RECEPTOR, ... | | Authors: | Szakonyi, G, Guthridge, J.M, Li, D, Holers, V.M, Chen, X.S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-06-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of complement receptor 2 in complex with its C3d ligand.
Science, 292, 2001
|
|
2R5K
 
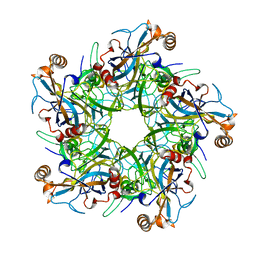 | |
2R5J
 
 | |
8EDJ
 
 | | Crystal structure of rA3G-ssRNA-GA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*GP*AP*UP*UP*U)-3'), SULFATE ION, ... | | Authors: | Pacheco, J, Yang, H.J, Li, S.-X, Chen, X.S. | | Deposit date: | 2022-09-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
6P3Y
 
 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 7.4) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
6KZQ
 
 | | structure of PTP-MEG2 and NSF-pY83 peptide complex | | Descriptor: | NSF-pY83 peptide, Tyrosine-protein phosphatase non-receptor type 9 | | Authors: | Xu, Y.F, Chen, X, Yu, X, Sun, J.P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PTP-MEG2 regulates quantal size and fusion pore opening through two distinct structural bases and substrates.
Embo Rep., 22, 2021
|
|
6L03
 
 | | structure of PTP-MEG2 and MUNC18-1-pY145 peptide complex | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 9, stxbp1-pY145 peptide | | Authors: | Xu, Y.F, Chen, X, Yu, X, Sun, J.P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | PTP-MEG2 regulates quantal size and fusion pore opening through two distinct structural bases and substrates.
Embo Rep., 22, 2021
|
|
6P40
 
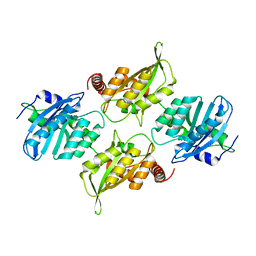 | | Crystal Structure of Full Length APOBEC3G FKL | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
