6PXQ
 
 | | Crystal structure of human thrombin mutant D194A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin heavy chain, Thrombin light chain | | Authors: | Stojanovski, B, Chen, Z, Koester, S.K, Pelc, L.A, Di Cera, E. | | Deposit date: | 2019-07-26 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Role of the I16-D194 ionic interaction in the trypsin fold.
Sci Rep, 9, 2019
|
|
2PGQ
 
 | | Human thrombin mutant C191A-C220A in complex with the inhibitor PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Thrombin heavy chain, ... | | Authors: | Bush-Pelc, L.A, Marino, F, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-04-10 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Important role of the cys-191 cys-220 disulfide bond in thrombin function and allostery
J.Biol.Chem., 282, 2007
|
|
2PGB
 
 | | Inhibitor-free human thrombin mutant C191A-C220A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin, SULFATE ION | | Authors: | Bush-Pelc, L.A, Marino, F, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-04-09 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Important role of the cys-191 cys-220 disulfide bond in thrombin function and allostery
J.Biol.Chem., 282, 2007
|
|
2PUX
 
 | | Crystal structure of murine thrombin in complex with the extracellular fragment of murine PAR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 3, Thrombin heavy chain, ... | | Authors: | Bah, A, Chen, Z, Bush-Pelc, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1WVE
 
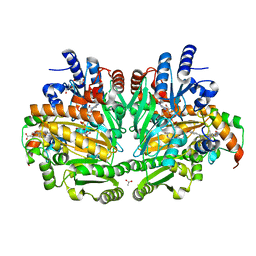 | | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon its Binding to the Cytochrome Subunit | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-cresol dehydrogenase [hydroxylating] cytochrome c subunit, 4-cresol dehydrogenase [hydroxylating] flavoprotein subunit, ... | | Authors: | Cunane, L.M, Chen, Z.-W, McIntire, W.S, Mathews, F.S. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon Its Binding to the Cytochrome Subunit
Biochemistry, 44, 2005
|
|
1L9E
 
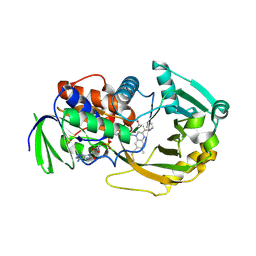 | | Role of Histidine 269 in Catalysis by Monomeric Sarcosine Oxidase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Zhao, G, Song, H, Chen, Z.-w, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-08-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Monomeric sarcosine oxidase: role of histidine 269 in catalysis.
Biochemistry, 41, 2002
|
|
1GG4
 
 | |
7SR9
 
 | | Human alpha-thrombin with 180- and 220- loops replaced with homologous loops from protein C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Di Cera, E, Ruben, E.A, Chen, Z. | | Deposit date: | 2021-11-08 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The active site region plays a critical role in Na + binding to thrombin.
J.Biol.Chem., 298, 2022
|
|
1L9C
 
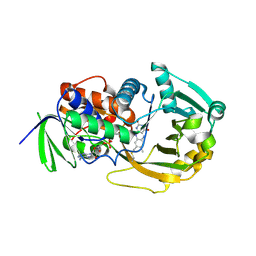 | | Role of Histidine 269 in Catalysis by Monomeric Sarcosine Oxidase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric Sarcosine Oxidase, ... | | Authors: | Zhao, G, Song, H, Chen, Z.-w, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-08-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Monomeric sarcosine oxidase: role of histidine 269 in catalysis.
Biochemistry, 41, 2002
|
|
1L9D
 
 | | Role of Histidine 269 in Catalysis by Monomeric Sarcosine Oxidase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase, ... | | Authors: | Zhao, G, Song, H, Chen, Z.-w, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-08-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Monomeric sarcosine oxidase: role of histidine 269 in catalysis.
Biochemistry, 41, 2002
|
|
6V5T
 
 | |
6V64
 
 | | Crystal structure of human thrombin bound to ppack with tryptophans replaced by 5-F-tryptophan | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Ruben, E.A, Chen, Z, Di Cera, E. | | Deposit date: | 2019-12-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | 19F NMR reveals the conformational properties of free thrombin and its zymogen precursor prethrombin-2.
J.Biol.Chem., 295, 2020
|
|
6A59
 
 | | Structure of histone demethylase REF6 at 1.8A | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
4GQH
 
 | | The Conformations and Interactions of the Four-Layer Aggregate Revealed by X-ray Crystallography Diffraction Implied the Importance of Peptides at Opposite Ends in Their Assemblies | | Descriptor: | Capsid protein | | Authors: | Li, X.Y, Song, B.A, Hu, D.Y, Chen, X, Wang, Z.C, Zeng, M.J, Yu, D.D, Chen, Z, Jin, L.H, Yang, S. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The Conformations and Interactions of the Four-Layer Aggregate Revealed by X-ray Crystallography Diffraction Implied the Importance of Peptides at Opposite Ends in Their Assemblies
To be Published
|
|
1JXP
 
 | | BK STRAIN HEPATITIS C VIRUS (HCV) NS3-NS4A | | Descriptor: | NS3 SERINE PROTEASE, NS4A, ZINC ION | | Authors: | Yan, Y, Munshi, S, Chen, Z. | | Deposit date: | 1997-08-21 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form.
Protein Sci., 7, 1998
|
|
7MOA
 
 | | Cryo-EM structure of the c-MET II/HGF I complex bound with HGF II in a rigid conformation | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MOB
 
 | | Cryo-EM structure of 2:2 c-MET/NK1 complex | | Descriptor: | Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO8
 
 | | Cryo-EM structure of 1:1 c-MET I/HGF I complex after focused 3D refinement of holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO7
 
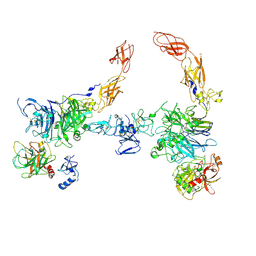 | | Cryo-EM structure of 2:2 c-MET/HGF holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO9
 
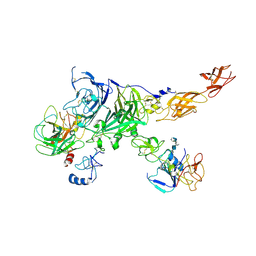 | | Cryo-EM map of the c-MET II/HGF I/HGF II (K4 and SPH) sub-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
5HC1
 
 | |
5HBZ
 
 | |
1NWO
 
 | |
8HAZ
 
 | |
8HBB
 
 | |
