8KGP
 
 | |
8KGS
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA topoisomerase 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGQ
 
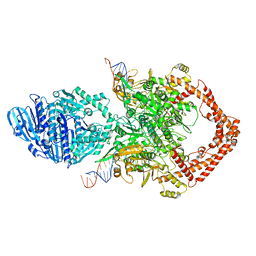 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (38-MER), DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGL
 
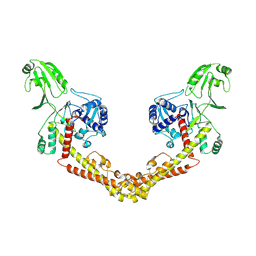 | |
8KGR
 
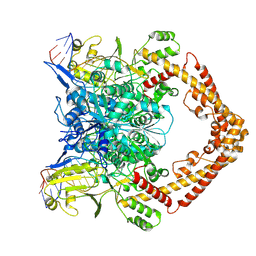 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (32-MER), DNA (33-MER), DNA topoisomerase 2, ... | | Authors: | Cong, J, Xin, U, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGM
 
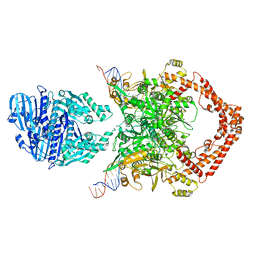 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (38-MER), DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGN
 
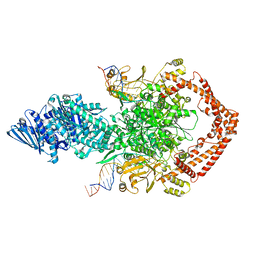 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (38-MER), DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
1SDI
 
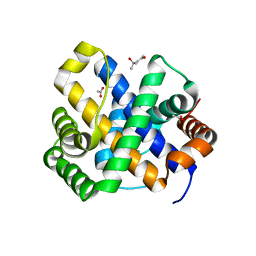 | | 1.65 A structure of Escherichia coli ycfC gene product | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Hypothetical protein ycfC | | Authors: | Borek, D, Otwinowski, Z, Chen, Y, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of Escherichia coli ycfC gene product
To be Published
|
|
1WPB
 
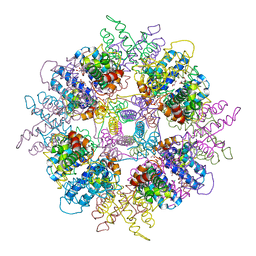 | | Structure of Escherichia coli yfbU gene product | | Descriptor: | CHLORIDE ION, GLYCEROL, hypothetical protein yfbU | | Authors: | Borek, D, Chen, Y, Zheng, M, Skarina, T, Savchenko, A, Edwards, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli yfbU gene product
To be Published
|
|
8D1P
 
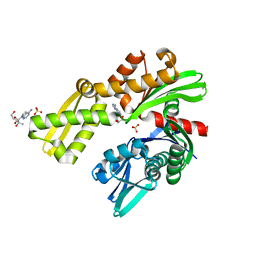 | | Crystal structure of Plasmodium falciparum GRP78-NBD in complex with 7-Deaza-2'-C-methyladenosine | | Descriptor: | 7-(2-C-methyl-beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
8D1S
 
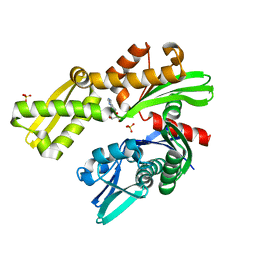 | | Crystal structure of Plasmodium falciparum GRP78 in complex with Toyocamycin | | Descriptor: | 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
8D1Q
 
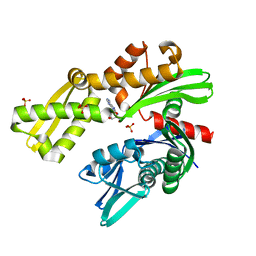 | | Crystal structure of Plasmodium falciparum GRP78-NBD in complex with 8-Aminoadenosine | | Descriptor: | (2R,3R,4S,5R)-2-(6,8-diaminopurin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
4UA9
 
 | |
4UAA
 
 | |
4UA6
 
 | |
4UA7
 
 | |
4E13
 
 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
4DYD
 
 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
4V8U
 
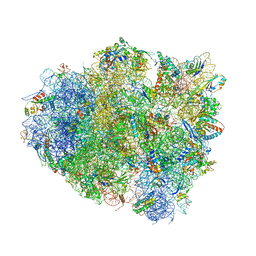 | | Crystal Structure of 70S Ribosome with Both Cognate tRNAs in the E and P Sites Representing an Authentic Elongation Complex. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, Y.G, Feng, S, Chen, Y. | | Deposit date: | 2012-08-28 | | Release date: | 2014-07-09 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex.
PLoS ONE, 8, 2013
|
|
4E12
 
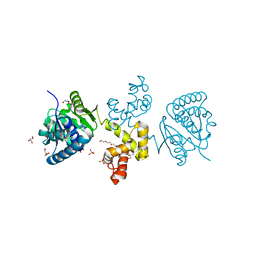 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
7BWQ
 
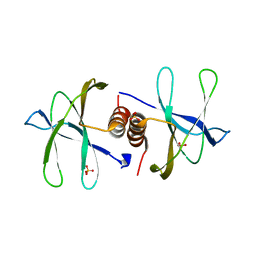 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | Descriptor: | Nsp9, SULFATE ION | | Authors: | Zhang, C, Chen, Y, Li, L, Su, D. | | Deposit date: | 2020-04-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
5K5T
 
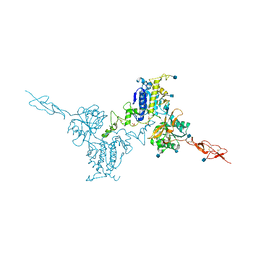 | | Crystal structure of the inactive form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
3WH9
 
 | | The ligand-free structure of ManBK from Aspergillus niger BK01 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, J.W, Chen, C.C, Huang, C.H, Huang, T.Y, Wu, T.H, Cheng, Y.S, Ko, T.P, Lin, C.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Analysis and Rational Design to Improve Specific Activity of beta-Mannanase from Aspergillus Niger BK01
To be Published
|
|
3WRE
 
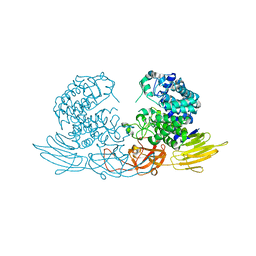 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRF
 
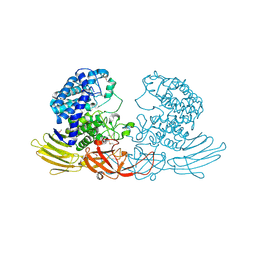 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
